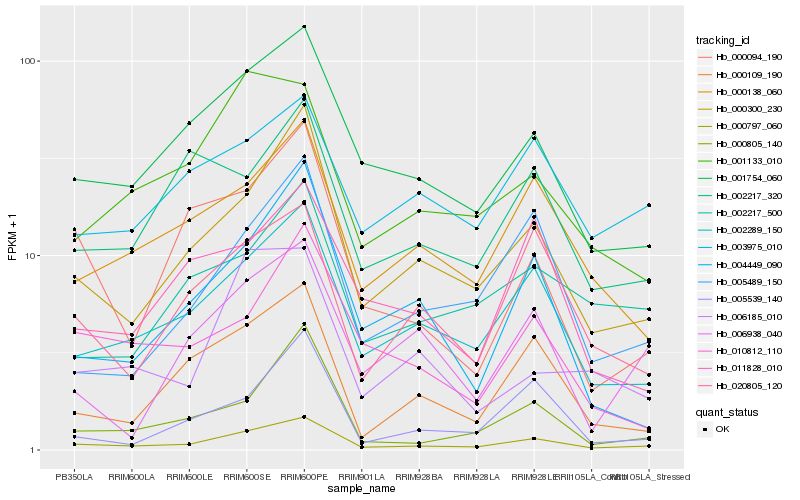
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000797_060 |
0.0 |
- |
- |
PREDICTED: subtilisin-like protease SBT1.7 [Jatropha curcas] |
| 2 |
Hb_000300_230 |
0.1032957744 |
- |
- |
PREDICTED: cation/calcium exchanger 2 [Jatropha curcas] |
| 3 |
Hb_002289_150 |
0.1128111415 |
- |
- |
PREDICTED: cellulose synthase A catalytic subunit 3 [UDP-forming]-like [Jatropha curcas] |
| 4 |
Hb_001754_060 |
0.1268365244 |
- |
- |
Xyloglucan 6-xylosyltransferase, putative [Ricinus communis] |
| 5 |
Hb_000109_190 |
0.1508758308 |
- |
- |
PREDICTED: RING-H2 finger protein ATL47 [Jatropha curcas] |
| 6 |
Hb_000138_060 |
0.1537856459 |
- |
- |
PREDICTED: protein trichome birefringence [Jatropha curcas] |
| 7 |
Hb_005489_150 |
0.1572354977 |
- |
- |
hypothetical protein JCGZ_20759 [Jatropha curcas] |
| 8 |
Hb_000805_140 |
0.1616188657 |
transcription factor |
TF Family: GRAS |
Chitin-inducible gibberellin-responsive protein, putative [Ricinus communis] |
| 9 |
Hb_020805_120 |
0.1654550413 |
- |
- |
PREDICTED: glutamate receptor 3.3 [Jatropha curcas] |
| 10 |
Hb_004449_090 |
0.1690777909 |
- |
- |
PREDICTED: CO(2)-response secreted protease-like [Jatropha curcas] |
| 11 |
Hb_002217_500 |
0.1778637594 |
- |
- |
hypothetical protein JCGZ_19014 [Jatropha curcas] |
| 12 |
Hb_001133_010 |
0.1789365896 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_000094_190 |
0.1789469583 |
- |
- |
PREDICTED: glucan endo-1,3-beta-glucosidase 7-like [Jatropha curcas] |
| 14 |
Hb_005539_140 |
0.1795258255 |
- |
- |
PREDICTED: U-box domain-containing protein 19-like [Jatropha curcas] |
| 15 |
Hb_010812_110 |
0.1821591512 |
- |
- |
putative plant disease resistance family protein [Populus trichocarpa] |
| 16 |
Hb_003975_010 |
0.1831493792 |
- |
- |
PREDICTED: protein RTF2 homolog [Jatropha curcas] |
| 17 |
Hb_002217_320 |
0.1844723105 |
- |
- |
PREDICTED: probable O-acetyltransferase CAS1 isoform X2 [Jatropha curcas] |
| 18 |
Hb_006185_010 |
0.1845307132 |
- |
- |
hypothetical protein L484_013453 [Morus notabilis] |
| 19 |
Hb_006938_040 |
0.1847653453 |
- |
- |
PREDICTED: uncharacterized protein At1g66480-like [Populus euphratica] |
| 20 |
Hb_011828_010 |
0.1849741503 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At1g67720 [Jatropha curcas] |