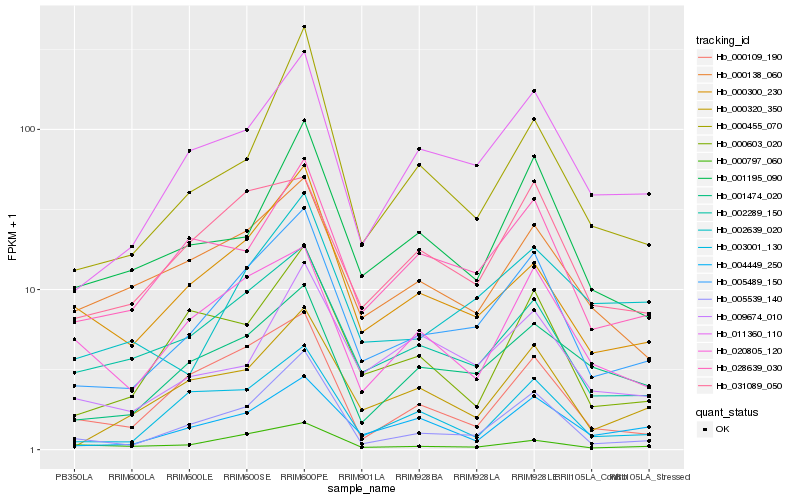
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005489_150 |
0.0 |
- |
- |
hypothetical protein JCGZ_20759 [Jatropha curcas] |
| 2 |
Hb_005539_140 |
0.1161032688 |
- |
- |
PREDICTED: U-box domain-containing protein 19-like [Jatropha curcas] |
| 3 |
Hb_011360_110 |
0.1164520919 |
- |
- |
PREDICTED: UDP-glucose 6-dehydrogenase 1 [Vitis vinifera] |
| 4 |
Hb_002289_150 |
0.1293201351 |
- |
- |
PREDICTED: cellulose synthase A catalytic subunit 3 [UDP-forming]-like [Jatropha curcas] |
| 5 |
Hb_028639_030 |
0.1417350429 |
- |
- |
PREDICTED: transmembrane 9 superfamily member 2-like [Jatropha curcas] |
| 6 |
Hb_000320_350 |
0.1422839934 |
- |
- |
hypothetical protein RCOM_0904330 [Ricinus communis] |
| 7 |
Hb_000300_230 |
0.144855476 |
- |
- |
PREDICTED: cation/calcium exchanger 2 [Jatropha curcas] |
| 8 |
Hb_001474_020 |
0.1470246094 |
- |
- |
PREDICTED: uncharacterized protein LOC105650917 [Jatropha curcas] |
| 9 |
Hb_004449_250 |
0.1522711683 |
- |
- |
PREDICTED: protein EI24 homolog [Jatropha curcas] |
| 10 |
Hb_001195_090 |
0.1533096571 |
- |
- |
PREDICTED: isoflavone reductase-like protein [Jatropha curcas] |
| 11 |
Hb_031089_050 |
0.1551817893 |
- |
- |
kinase, putative [Ricinus communis] |
| 12 |
Hb_000797_060 |
0.1572354977 |
- |
- |
PREDICTED: subtilisin-like protease SBT1.7 [Jatropha curcas] |
| 13 |
Hb_003001_130 |
0.157962904 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_000109_190 |
0.1608652995 |
- |
- |
PREDICTED: RING-H2 finger protein ATL47 [Jatropha curcas] |
| 15 |
Hb_000138_060 |
0.1623557682 |
- |
- |
PREDICTED: protein trichome birefringence [Jatropha curcas] |
| 16 |
Hb_002639_020 |
0.1628967726 |
- |
- |
PREDICTED: probable alpha-amylase 2 [Jatropha curcas] |
| 17 |
Hb_020805_120 |
0.1637208148 |
- |
- |
PREDICTED: glutamate receptor 3.3 [Jatropha curcas] |
| 18 |
Hb_000455_070 |
0.1650793666 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: carboxymethylenebutenolidase homolog [Jatropha curcas] |
| 19 |
Hb_000603_020 |
0.1667309926 |
- |
- |
PREDICTED: uncharacterized protein LOC105638063 isoform X1 [Jatropha curcas] |
| 20 |
Hb_009674_010 |
0.1667382319 |
- |
- |
AAEL007687-PA [Aedes aegypti] |