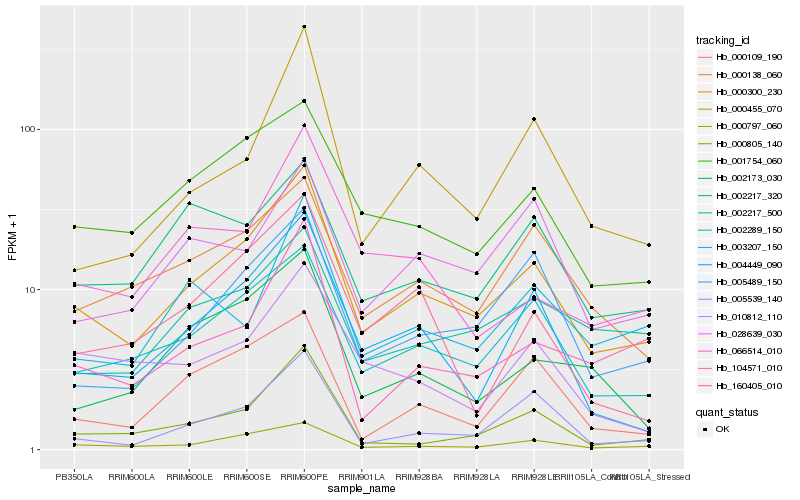
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000300_230 |
0.0 |
- |
- |
PREDICTED: cation/calcium exchanger 2 [Jatropha curcas] |
| 2 |
Hb_000797_060 |
0.1032957744 |
- |
- |
PREDICTED: subtilisin-like protease SBT1.7 [Jatropha curcas] |
| 3 |
Hb_002289_150 |
0.10801239 |
- |
- |
PREDICTED: cellulose synthase A catalytic subunit 3 [UDP-forming]-like [Jatropha curcas] |
| 4 |
Hb_001754_060 |
0.129169276 |
- |
- |
Xyloglucan 6-xylosyltransferase, putative [Ricinus communis] |
| 5 |
Hb_004449_090 |
0.1348098129 |
- |
- |
PREDICTED: CO(2)-response secreted protease-like [Jatropha curcas] |
| 6 |
Hb_000138_060 |
0.1440287683 |
- |
- |
PREDICTED: protein trichome birefringence [Jatropha curcas] |
| 7 |
Hb_005489_150 |
0.144855476 |
- |
- |
hypothetical protein JCGZ_20759 [Jatropha curcas] |
| 8 |
Hb_000805_140 |
0.1528511564 |
transcription factor |
TF Family: GRAS |
Chitin-inducible gibberellin-responsive protein, putative [Ricinus communis] |
| 9 |
Hb_002217_500 |
0.1536170383 |
- |
- |
hypothetical protein JCGZ_19014 [Jatropha curcas] |
| 10 |
Hb_005539_140 |
0.1547046299 |
- |
- |
PREDICTED: U-box domain-containing protein 19-like [Jatropha curcas] |
| 11 |
Hb_000109_190 |
0.1558952085 |
- |
- |
PREDICTED: RING-H2 finger protein ATL47 [Jatropha curcas] |
| 12 |
Hb_000455_070 |
0.1564940342 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: carboxymethylenebutenolidase homolog [Jatropha curcas] |
| 13 |
Hb_104571_010 |
0.1582690233 |
- |
- |
PREDICTED: metal transporter Nramp1 [Jatropha curcas] |
| 14 |
Hb_028639_030 |
0.161940897 |
- |
- |
PREDICTED: transmembrane 9 superfamily member 2-like [Jatropha curcas] |
| 15 |
Hb_002173_030 |
0.1622732908 |
- |
- |
hypothetical protein JCGZ_12466 [Jatropha curcas] |
| 16 |
Hb_002217_320 |
0.1643013739 |
- |
- |
PREDICTED: probable O-acetyltransferase CAS1 isoform X2 [Jatropha curcas] |
| 17 |
Hb_003207_150 |
0.1648829645 |
- |
- |
PREDICTED: SPX domain-containing membrane protein At4g22990-like [Jatropha curcas] |
| 18 |
Hb_066514_010 |
0.1662339932 |
- |
- |
hypothetical protein RCOM_0204720 [Ricinus communis] |
| 19 |
Hb_010812_110 |
0.166718579 |
- |
- |
putative plant disease resistance family protein [Populus trichocarpa] |
| 20 |
Hb_160405_010 |
0.1671015968 |
- |
- |
NBS-LRR resistance protein RGH1 [Manihot esculenta] |