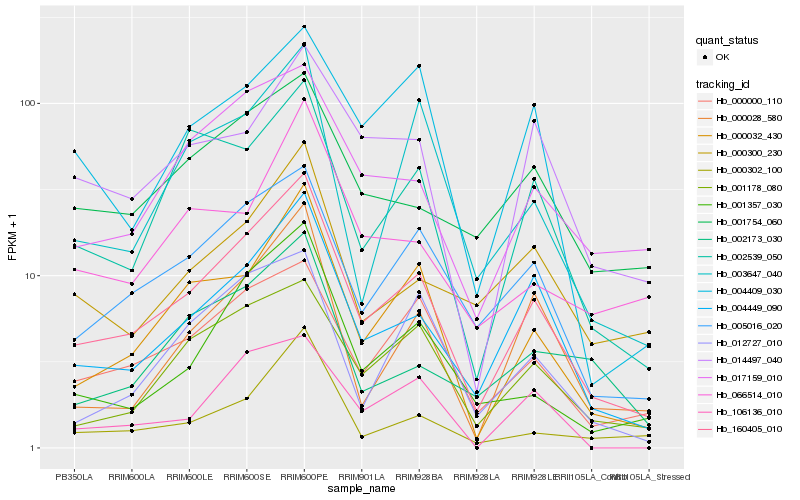
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_160405_010 |
0.0 |
- |
- |
NBS-LRR resistance protein RGH1 [Manihot esculenta] |
| 2 |
Hb_004449_090 |
0.1107960908 |
- |
- |
PREDICTED: CO(2)-response secreted protease-like [Jatropha curcas] |
| 3 |
Hb_000032_430 |
0.1170952253 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_005016_020 |
0.1223146106 |
desease resistance |
Gene Name: ABC_membrane |
ABC transporter family protein [Hevea brasiliensis] |
| 5 |
Hb_000000_110 |
0.1381446383 |
- |
- |
hypothetical protein CISIN_1g0027772mg, partial [Citrus sinensis] |
| 6 |
Hb_001357_030 |
0.144179975 |
- |
- |
hypothetical protein F383_04880 [Gossypium arboreum] |
| 7 |
Hb_001178_080 |
0.1463970229 |
- |
- |
PREDICTED: protein kinase PVPK-1 [Jatropha curcas] |
| 8 |
Hb_017159_010 |
0.1476237231 |
- |
- |
- |
| 9 |
Hb_002539_050 |
0.1545608074 |
- |
- |
PREDICTED: PLASMODESMATA CALLOSE-BINDING PROTEIN 3 [Jatropha curcas] |
| 10 |
Hb_000028_580 |
0.15773208 |
- |
- |
PREDICTED: glutamate decarboxylase 5-like [Jatropha curcas] |
| 11 |
Hb_003647_040 |
0.1599699696 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_000302_100 |
0.1640840259 |
- |
- |
PREDICTED: uncharacterized protein LOC105766209 [Gossypium raimondii] |
| 13 |
Hb_000300_230 |
0.1671015968 |
- |
- |
PREDICTED: cation/calcium exchanger 2 [Jatropha curcas] |
| 14 |
Hb_001754_060 |
0.1671594772 |
- |
- |
Xyloglucan 6-xylosyltransferase, putative [Ricinus communis] |
| 15 |
Hb_012727_010 |
0.1676842194 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: disease resistance RPP13-like protein 4 [Jatropha curcas] |
| 16 |
Hb_014497_040 |
0.1687685177 |
- |
- |
PREDICTED: calvin cycle protein CP12-3, chloroplastic [Jatropha curcas] |
| 17 |
Hb_106136_010 |
0.172103705 |
- |
- |
Disease resistance protein RPS5, putative [Ricinus communis] |
| 18 |
Hb_002173_030 |
0.1727532306 |
- |
- |
hypothetical protein JCGZ_12466 [Jatropha curcas] |
| 19 |
Hb_066514_010 |
0.17511869 |
- |
- |
hypothetical protein RCOM_0204720 [Ricinus communis] |
| 20 |
Hb_004409_030 |
0.1771711108 |
- |
- |
Glutamine synthetase 1,4 [Theobroma cacao] |