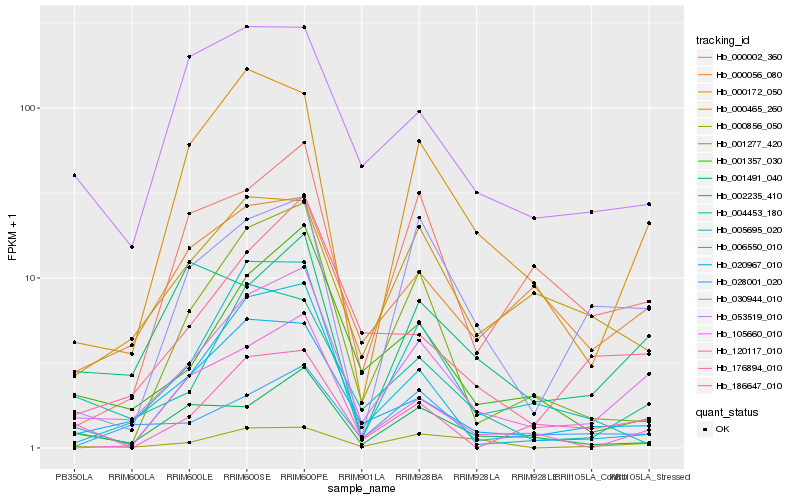
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_105660_010 |
0.0 |
- |
- |
PREDICTED: potassium channel AKT6-like [Jatropha curcas] |
| 2 |
Hb_020967_010 |
0.1602333215 |
- |
- |
- |
| 3 |
Hb_004453_180 |
0.1639224384 |
- |
- |
hypothetical protein JCGZ_07188 [Jatropha curcas] |
| 4 |
Hb_001357_030 |
0.172001865 |
- |
- |
hypothetical protein F383_04880 [Gossypium arboreum] |
| 5 |
Hb_186647_010 |
0.1767370483 |
- |
- |
- |
| 6 |
Hb_000172_050 |
0.1854120214 |
- |
- |
PREDICTED: uncharacterized protein LOC105650611 [Jatropha curcas] |
| 7 |
Hb_006550_010 |
0.1883437863 |
- |
- |
PREDICTED: vacuolar amino acid transporter 1 [Eucalyptus grandis] |
| 8 |
Hb_000056_080 |
0.1926406071 |
- |
- |
PREDICTED: protein kinase 2B, chloroplastic [Jatropha curcas] |
| 9 |
Hb_001277_420 |
0.1926565155 |
- |
- |
PREDICTED: protein lin-12-like isoform X2 [Populus euphratica] |
| 10 |
Hb_028001_020 |
0.1960933464 |
- |
- |
PREDICTED: lysM domain receptor-like kinase 4 [Jatropha curcas] |
| 11 |
Hb_120117_010 |
0.1980631838 |
- |
- |
Disease resistance protein RFL1, putative [Ricinus communis] |
| 12 |
Hb_000856_050 |
0.1990544019 |
- |
- |
PREDICTED: HIPL1 protein-like [Jatropha curcas] |
| 13 |
Hb_053519_010 |
0.1996101849 |
- |
- |
PREDICTED: probable zinc transporter protein DDB_G0282067 [Jatropha curcas] |
| 14 |
Hb_176894_010 |
0.1996951119 |
- |
- |
- |
| 15 |
Hb_000002_360 |
0.2003896813 |
- |
- |
hypothetical protein POPTR_0008s16470g [Populus trichocarpa] |
| 16 |
Hb_001491_040 |
0.2042327074 |
- |
- |
PREDICTED: probable leucine-rich repeat receptor-like protein kinase At1g35710 [Jatropha curcas] |
| 17 |
Hb_030944_010 |
0.2047537484 |
- |
- |
PREDICTED: uncharacterized protein LOC105633455 [Jatropha curcas] |
| 18 |
Hb_000465_260 |
0.2122949353 |
transcription factor |
TF Family: HB |
PREDICTED: homeobox-leucine zipper protein HAT4-like [Jatropha curcas] |
| 19 |
Hb_002235_410 |
0.2126981967 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RNF181-like [Jatropha curcas] |
| 20 |
Hb_005695_020 |
0.2129281175 |
- |
- |
PREDICTED: low affinity sulfate transporter 3 [Jatropha curcas] |