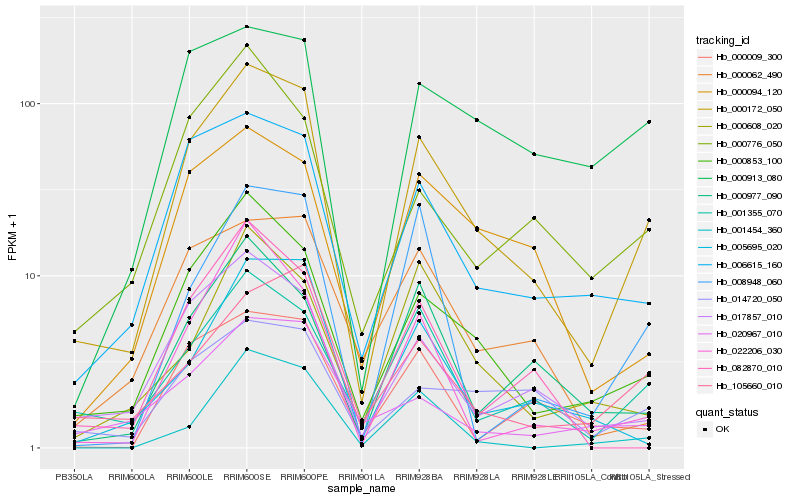
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000172_050 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105650611 [Jatropha curcas] |
| 2 |
Hb_000853_100 |
0.1199800433 |
transcription factor |
TF Family: ERF |
PREDICTED: ethylene-responsive transcription factor ERF073 [Jatropha curcas] |
| 3 |
Hb_006615_160 |
0.1517218788 |
- |
- |
PREDICTED: UDP-glycosyltransferase 89B1-like [Jatropha curcas] |
| 4 |
Hb_001355_070 |
0.1526504351 |
- |
- |
P-loop containing nucleoside triphosphate hydrolases superfamily protein, putative isoform 2 [Theobroma cacao] |
| 5 |
Hb_017857_010 |
0.1546337356 |
- |
- |
- |
| 6 |
Hb_014720_050 |
0.1752958606 |
- |
- |
PREDICTED: serine/threonine-protein kinase ATM-like [Jatropha curcas] |
| 7 |
Hb_000094_120 |
0.1783284251 |
- |
- |
PREDICTED: uncharacterized protein At1g01500 [Jatropha curcas] |
| 8 |
Hb_000977_090 |
0.184479018 |
- |
- |
hypothetical protein POPTR_0010s18550g [Populus trichocarpa] |
| 9 |
Hb_105660_010 |
0.1854120214 |
- |
- |
PREDICTED: potassium channel AKT6-like [Jatropha curcas] |
| 10 |
Hb_000776_050 |
0.1864138545 |
- |
- |
Alpha-1,4-glucan-protein synthase [UDP-forming], putative [Ricinus communis] |
| 11 |
Hb_000009_300 |
0.1871191857 |
- |
- |
hypothetical protein CICLE_v10024731mg [Citrus clementina] |
| 12 |
Hb_000913_080 |
0.1916424749 |
- |
- |
PREDICTED: CBL-interacting serine/threonine-protein kinase 11 [Jatropha curcas] |
| 13 |
Hb_022206_030 |
0.1938280015 |
- |
- |
Auxin-induced in root cultures protein 12 precursor, putative [Ricinus communis] |
| 14 |
Hb_005695_020 |
0.1956580092 |
- |
- |
PREDICTED: low affinity sulfate transporter 3 [Jatropha curcas] |
| 15 |
Hb_020967_010 |
0.1963634264 |
- |
- |
- |
| 16 |
Hb_082870_010 |
0.1964034424 |
- |
- |
Disease resistance protein RFL1, putative [Ricinus communis] |
| 17 |
Hb_000062_490 |
0.199079274 |
- |
- |
PREDICTED: lipoxygenase 6, chloroplastic [Jatropha curcas] |
| 18 |
Hb_001454_360 |
0.200499459 |
- |
- |
PREDICTED: bidirectional sugar transporter SWEET17 [Jatropha curcas] |
| 19 |
Hb_008948_060 |
0.2009317122 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_000608_020 |
0.2014242371 |
- |
- |
PREDICTED: wall-associated receptor kinase-like 22 [Jatropha curcas] |