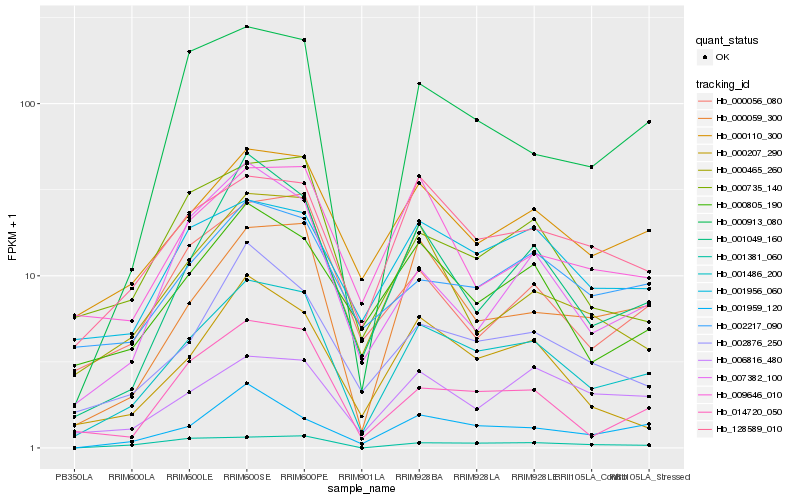
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001486_200 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105632606 [Jatropha curcas] |
| 2 |
Hb_000059_300 |
0.1261342315 |
- |
- |
caspase, putative [Ricinus communis] |
| 3 |
Hb_000110_300 |
0.1272473273 |
- |
- |
PREDICTED: uncharacterized protein LOC105642013 [Jatropha curcas] |
| 4 |
Hb_001049_160 |
0.1349033283 |
- |
- |
aldose 1-epimerase, putative [Ricinus communis] |
| 5 |
Hb_002876_250 |
0.1373631735 |
- |
- |
PREDICTED: receptor-like protein kinase 2 [Jatropha curcas] |
| 6 |
Hb_014720_050 |
0.1390050785 |
- |
- |
PREDICTED: serine/threonine-protein kinase ATM-like [Jatropha curcas] |
| 7 |
Hb_128589_010 |
0.1390735819 |
- |
- |
PREDICTED: cyclin-dependent kinase inhibitor 5 [Jatropha curcas] |
| 8 |
Hb_000207_290 |
0.1440006064 |
- |
- |
hypothetical protein JCGZ_03920 [Jatropha curcas] |
| 9 |
Hb_000805_190 |
0.1441863034 |
transcription factor |
TF Family: GRAS |
PREDICTED: uncharacterized protein LOC103323607 [Prunus mume] |
| 10 |
Hb_000735_140 |
0.146442333 |
- |
- |
3-glucanase family protein [Populus trichocarpa] |
| 11 |
Hb_000913_080 |
0.1476574308 |
- |
- |
PREDICTED: CBL-interacting serine/threonine-protein kinase 11 [Jatropha curcas] |
| 12 |
Hb_001956_060 |
0.1477480959 |
- |
- |
PREDICTED: oxysterol-binding protein-related protein 3C-like [Jatropha curcas] |
| 13 |
Hb_000465_260 |
0.1484824279 |
transcription factor |
TF Family: HB |
PREDICTED: homeobox-leucine zipper protein HAT4-like [Jatropha curcas] |
| 14 |
Hb_002217_090 |
0.1528922963 |
- |
- |
PREDICTED: probable protein phosphatase 2C 13 [Jatropha curcas] |
| 15 |
Hb_001959_120 |
0.1529164529 |
- |
- |
PREDICTED: uncharacterized protein LOC105638487 [Jatropha curcas] |
| 16 |
Hb_001381_060 |
0.1542128674 |
- |
- |
monoxygenase, putative [Ricinus communis] |
| 17 |
Hb_006816_480 |
0.1569161489 |
- |
- |
PREDICTED: F-box/LRR-repeat protein 3 isoform X1 [Jatropha curcas] |
| 18 |
Hb_000056_080 |
0.1571467746 |
- |
- |
PREDICTED: protein kinase 2B, chloroplastic [Jatropha curcas] |
| 19 |
Hb_007382_100 |
0.1598124931 |
- |
- |
receptor serine-threonine protein kinase, putative [Ricinus communis] |
| 20 |
Hb_009646_010 |
0.1612391989 |
- |
- |
PREDICTED: hyoscyamine 6-dioxygenase-like [Jatropha curcas] |