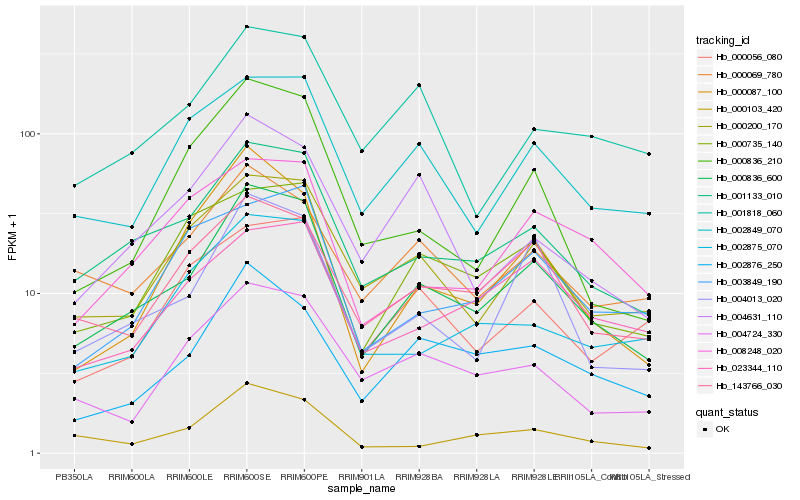
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000836_600 |
0.0 |
- |
- |
PREDICTED: delta(8)-fatty-acid desaturase 2 [Jatropha curcas] |
| 2 |
Hb_001133_010 |
0.080075236 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_004013_020 |
0.1181985486 |
- |
- |
PREDICTED: F-box protein SKIP2 [Jatropha curcas] |
| 4 |
Hb_000200_170 |
0.1189532435 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RHA2A-like [Populus euphratica] |
| 5 |
Hb_143766_030 |
0.1213220729 |
- |
- |
protein with unknown function [Ricinus communis] |
| 6 |
Hb_002876_250 |
0.1268499371 |
- |
- |
PREDICTED: receptor-like protein kinase 2 [Jatropha curcas] |
| 7 |
Hb_002849_070 |
0.128845491 |
- |
- |
PREDICTED: protein BPS1, chloroplastic [Jatropha curcas] |
| 8 |
Hb_000735_140 |
0.1309254545 |
- |
- |
3-glucanase family protein [Populus trichocarpa] |
| 9 |
Hb_000087_100 |
0.1372044043 |
- |
- |
PREDICTED: cellulose synthase-like protein D3 [Jatropha curcas] |
| 10 |
Hb_004724_330 |
0.1377753256 |
- |
- |
Capsanthin/capsorubin synthase, chloroplast precursor, putative [Ricinus communis] |
| 11 |
Hb_000069_780 |
0.1387729349 |
- |
- |
atpob1, putative [Ricinus communis] |
| 12 |
Hb_000103_420 |
0.1406136911 |
- |
- |
PREDICTED: putative calcium-transporting ATPase 13, plasma membrane-type [Populus euphratica] |
| 13 |
Hb_008248_020 |
0.1417672545 |
transcription factor |
TF Family: DBB |
zinc finger protein, putative [Ricinus communis] |
| 14 |
Hb_023344_110 |
0.1444124685 |
- |
- |
PREDICTED: xyloglucan galactosyltransferase KATAMARI1 [Jatropha curcas] |
| 15 |
Hb_002875_070 |
0.1461694764 |
- |
- |
PREDICTED: probable mitochondrial chaperone BCS1-A [Jatropha curcas] |
| 16 |
Hb_001818_060 |
0.1470017081 |
- |
- |
PREDICTED: cysteine proteinase inhibitor 5-like [Jatropha curcas] |
| 17 |
Hb_004631_110 |
0.1482969541 |
- |
- |
ring finger protein, putative [Ricinus communis] |
| 18 |
Hb_000056_080 |
0.1488102442 |
- |
- |
PREDICTED: protein kinase 2B, chloroplastic [Jatropha curcas] |
| 19 |
Hb_000836_210 |
0.1525404535 |
- |
- |
hypothetical protein JCGZ_14362 [Jatropha curcas] |
| 20 |
Hb_003849_190 |
0.1556353678 |
- |
- |
PREDICTED: uncharacterized protein LOC105644700 [Jatropha curcas] |