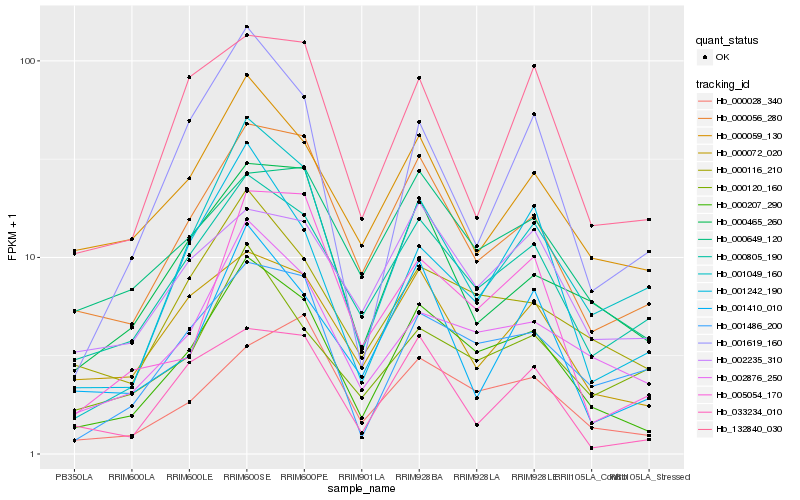
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000207_290 |
0.0 |
- |
- |
hypothetical protein JCGZ_03920 [Jatropha curcas] |
| 2 |
Hb_000805_190 |
0.1210254703 |
transcription factor |
TF Family: GRAS |
PREDICTED: uncharacterized protein LOC103323607 [Prunus mume] |
| 3 |
Hb_002876_250 |
0.1216091193 |
- |
- |
PREDICTED: receptor-like protein kinase 2 [Jatropha curcas] |
| 4 |
Hb_000056_280 |
0.1254073411 |
- |
- |
sur2 hydroxylase/desaturase, putative [Ricinus communis] |
| 5 |
Hb_000116_210 |
0.1260631848 |
- |
- |
PREDICTED: uncharacterized protein LOC105628567 [Jatropha curcas] |
| 6 |
Hb_001049_160 |
0.1407748081 |
- |
- |
aldose 1-epimerase, putative [Ricinus communis] |
| 7 |
Hb_001486_200 |
0.1440006064 |
- |
- |
PREDICTED: uncharacterized protein LOC105632606 [Jatropha curcas] |
| 8 |
Hb_000072_020 |
0.147494028 |
- |
- |
PREDICTED: C2 and GRAM domain-containing protein At1g03370 isoform X2 [Jatropha curcas] |
| 9 |
Hb_001242_190 |
0.1491359034 |
- |
- |
PREDICTED: type I inositol 1,4,5-trisphosphate 5-phosphatase 1 isoform X3 [Jatropha curcas] |
| 10 |
Hb_000059_130 |
0.149518425 |
transcription factor |
TF Family: GRAS |
PREDICTED: scarecrow-like protein 1 [Jatropha curcas] |
| 11 |
Hb_000465_260 |
0.1533676329 |
transcription factor |
TF Family: HB |
PREDICTED: homeobox-leucine zipper protein HAT4-like [Jatropha curcas] |
| 12 |
Hb_001619_160 |
0.161462672 |
transcription factor |
TF Family: ERF |
DREB1p [Hevea brasiliensis] |
| 13 |
Hb_000649_120 |
0.1625569209 |
- |
- |
ankyrin repeat-containing protein, putative [Ricinus communis] |
| 14 |
Hb_000028_340 |
0.1647397256 |
- |
- |
PREDICTED: potassium channel SKOR-like [Prunus mume] |
| 15 |
Hb_002235_310 |
0.1649891999 |
- |
- |
PREDICTED: KH domain-containing protein At4g18375 [Jatropha curcas] |
| 16 |
Hb_000120_160 |
0.1662616661 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 17 |
Hb_033234_010 |
0.1673764402 |
desease resistance |
Gene Name: LRR_8 |
Disease resistance protein RPP8 [Theobroma cacao] |
| 18 |
Hb_001410_010 |
0.1674007156 |
transcription factor |
TF Family: RWP-RK |
PREDICTED: protein NLP7-like [Jatropha curcas] |
| 19 |
Hb_132840_030 |
0.167633403 |
transcription factor |
TF Family: MYB-related |
lhy1 [Populus tremula] |
| 20 |
Hb_005054_170 |
0.1680890051 |
transcription factor |
TF Family: MYB |
DNA binding protein, putative [Ricinus communis] |