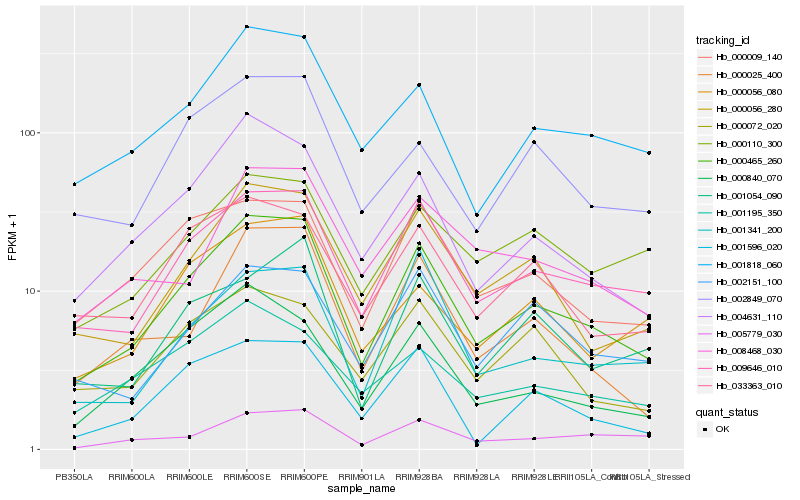
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000465_260 |
0.0 |
transcription factor |
TF Family: HB |
PREDICTED: homeobox-leucine zipper protein HAT4-like [Jatropha curcas] |
| 2 |
Hb_009646_010 |
0.0846432578 |
- |
- |
PREDICTED: hyoscyamine 6-dioxygenase-like [Jatropha curcas] |
| 3 |
Hb_000056_280 |
0.1062372198 |
- |
- |
sur2 hydroxylase/desaturase, putative [Ricinus communis] |
| 4 |
Hb_004631_110 |
0.1098839562 |
- |
- |
ring finger protein, putative [Ricinus communis] |
| 5 |
Hb_001818_060 |
0.1153191384 |
- |
- |
PREDICTED: cysteine proteinase inhibitor 5-like [Jatropha curcas] |
| 6 |
Hb_000025_400 |
0.1224699998 |
- |
- |
PREDICTED: uncharacterized protein LOC105110770 [Populus euphratica] |
| 7 |
Hb_033363_010 |
0.1245032625 |
transcription factor |
TF Family: NAC |
PREDICTED: NAC domain-containing protein 8 [Jatropha curcas] |
| 8 |
Hb_000009_140 |
0.1261393725 |
transcription factor |
TF Family: BES1 |
BRASSINAZOLE-RESISTANT 1 protein, putative [Ricinus communis] |
| 9 |
Hb_002849_070 |
0.1284922949 |
- |
- |
PREDICTED: protein BPS1, chloroplastic [Jatropha curcas] |
| 10 |
Hb_000840_070 |
0.1311527196 |
desease resistance |
Gene Name: NB-ARC |
leucine-rich repeat-containing protein, putative [Ricinus communis] |
| 11 |
Hb_000056_080 |
0.1315136877 |
- |
- |
PREDICTED: protein kinase 2B, chloroplastic [Jatropha curcas] |
| 12 |
Hb_001596_020 |
0.1317080708 |
- |
- |
hypothetical protein CICLE_v10014140mg [Citrus clementina] |
| 13 |
Hb_005779_030 |
0.1339193307 |
- |
- |
PREDICTED: nuclear pore complex protein NUP62 [Jatropha curcas] |
| 14 |
Hb_002151_100 |
0.1345619228 |
- |
- |
PREDICTED: uncharacterized protein LOC105637951 [Jatropha curcas] |
| 15 |
Hb_001054_090 |
0.137838143 |
- |
- |
Potassium channel SKOR, putative [Ricinus communis] |
| 16 |
Hb_001341_200 |
0.1398508274 |
transcription factor |
TF Family: ARF |
PREDICTED: auxin response factor 4 [Jatropha curcas] |
| 17 |
Hb_000110_300 |
0.1406776517 |
- |
- |
PREDICTED: uncharacterized protein LOC105642013 [Jatropha curcas] |
| 18 |
Hb_000072_020 |
0.1414460356 |
- |
- |
PREDICTED: C2 and GRAM domain-containing protein At1g03370 isoform X2 [Jatropha curcas] |
| 19 |
Hb_008468_030 |
0.1423370039 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: disease resistance RPP13-like protein 4 [Jatropha curcas] |
| 20 |
Hb_001195_350 |
0.1425881052 |
- |
- |
hypothetical protein JCGZ_04423 [Jatropha curcas] |