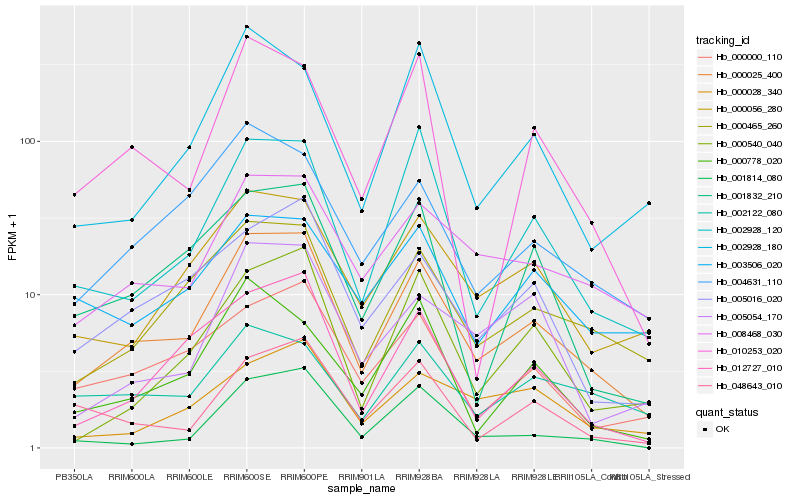
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000025_400 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105110770 [Populus euphratica] |
| 2 |
Hb_000465_260 |
0.1224699998 |
transcription factor |
TF Family: HB |
PREDICTED: homeobox-leucine zipper protein HAT4-like [Jatropha curcas] |
| 3 |
Hb_008468_030 |
0.1297081802 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: disease resistance RPP13-like protein 4 [Jatropha curcas] |
| 4 |
Hb_000056_280 |
0.1338935849 |
- |
- |
sur2 hydroxylase/desaturase, putative [Ricinus communis] |
| 5 |
Hb_002928_120 |
0.1385289002 |
- |
- |
1,4-alpha-glucan branching enzyme [Manihot esculenta] |
| 6 |
Hb_005016_020 |
0.141313151 |
desease resistance |
Gene Name: ABC_membrane |
ABC transporter family protein [Hevea brasiliensis] |
| 7 |
Hb_004631_110 |
0.1420103129 |
- |
- |
ring finger protein, putative [Ricinus communis] |
| 8 |
Hb_012727_010 |
0.142780514 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: disease resistance RPP13-like protein 4 [Jatropha curcas] |
| 9 |
Hb_000000_110 |
0.1453326049 |
- |
- |
hypothetical protein CISIN_1g0027772mg, partial [Citrus sinensis] |
| 10 |
Hb_000540_040 |
0.1518253293 |
- |
- |
PREDICTED: chitinase 2 [Amborella trichopoda] |
| 11 |
Hb_001832_210 |
0.1537982629 |
- |
- |
PREDICTED: venom phosphodiesterase 2 [Jatropha curcas] |
| 12 |
Hb_010253_020 |
0.1588347196 |
- |
- |
alcohol dehydrogenase, putative [Ricinus communis] |
| 13 |
Hb_002928_180 |
0.1606369934 |
- |
- |
catalase CAT1 [Manihot esculenta] |
| 14 |
Hb_048643_010 |
0.1614308619 |
- |
- |
NBS-LRR resistance protein RGH1 [Manihot esculenta] |
| 15 |
Hb_001814_080 |
0.1621384953 |
- |
- |
PREDICTED: oleosin 1-like [Jatropha curcas] |
| 16 |
Hb_000028_340 |
0.1632923474 |
- |
- |
PREDICTED: potassium channel SKOR-like [Prunus mume] |
| 17 |
Hb_005054_170 |
0.1635326626 |
transcription factor |
TF Family: MYB |
DNA binding protein, putative [Ricinus communis] |
| 18 |
Hb_000778_020 |
0.1666420715 |
- |
- |
PREDICTED: salicylic acid-binding protein 2-like [Jatropha curcas] |
| 19 |
Hb_003506_020 |
0.1682302191 |
- |
- |
PREDICTED: uncharacterized protein LOC105125881 isoform X1 [Populus euphratica] |
| 20 |
Hb_002122_080 |
0.1685744938 |
- |
- |
- |