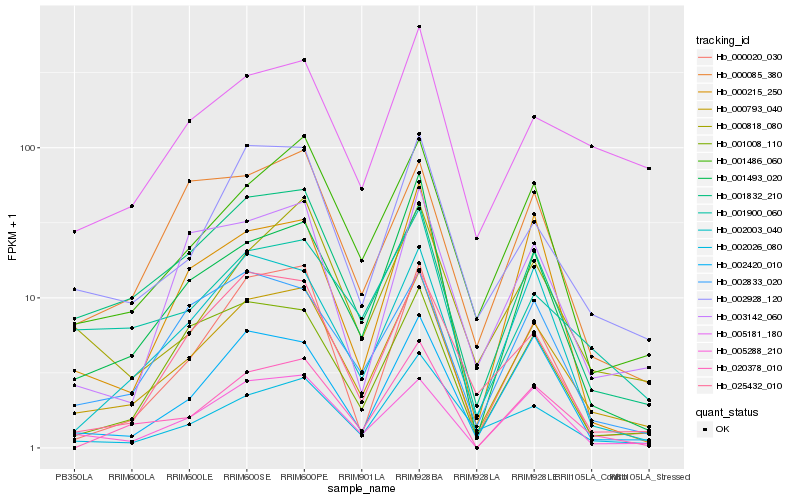
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000793_040 |
0.0 |
- |
- |
PREDICTED: transcription factor LHW [Jatropha curcas] |
| 2 |
Hb_001486_060 |
0.1093194869 |
- |
- |
PREDICTED: uncharacterized protein LOC105632622 [Jatropha curcas] |
| 3 |
Hb_002928_120 |
0.1105914859 |
- |
- |
1,4-alpha-glucan branching enzyme [Manihot esculenta] |
| 4 |
Hb_005288_210 |
0.1193891958 |
- |
- |
hypothetical protein VITISV_017217 [Vitis vinifera] |
| 5 |
Hb_020378_010 |
0.1208387978 |
- |
- |
PREDICTED: acyl-CoA--sterol O-acyltransferase 1 [Jatropha curcas] |
| 6 |
Hb_001493_020 |
0.1244288055 |
- |
- |
leucine-rich repeat receptor-like protein kinase [Manihot esculenta] |
| 7 |
Hb_002420_010 |
0.129373938 |
- |
- |
PREDICTED: uncharacterized protein LOC105124982 [Populus euphratica] |
| 8 |
Hb_002026_080 |
0.1306680374 |
desease resistance |
Gene Name: TIR_2 |
nucleoside-triphosphatase, putative [Ricinus communis] |
| 9 |
Hb_002003_040 |
0.1318679006 |
- |
- |
PREDICTED: probably inactive leucine-rich repeat receptor-like protein kinase At5g48380 isoform X1 [Jatropha curcas] |
| 10 |
Hb_001008_110 |
0.1331862606 |
- |
- |
- |
| 11 |
Hb_002833_020 |
0.1361966502 |
desease resistance |
Gene Name: LRR_4 |
PREDICTED: uncharacterized protein LOC105124982 [Populus euphratica] |
| 12 |
Hb_001900_060 |
0.1365939418 |
- |
- |
PREDICTED: neutral alpha-glucosidase C [Jatropha curcas] |
| 13 |
Hb_025432_010 |
0.1372198138 |
- |
- |
hypothetical protein JCGZ_21627 [Jatropha curcas] |
| 14 |
Hb_000818_080 |
0.1374596296 |
- |
- |
PREDICTED: monosaccharide-sensing protein 2 [Jatropha curcas] |
| 15 |
Hb_001832_210 |
0.1375392252 |
- |
- |
PREDICTED: venom phosphodiesterase 2 [Jatropha curcas] |
| 16 |
Hb_000020_030 |
0.1387444088 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 17 |
Hb_003142_060 |
0.1449365341 |
- |
- |
PREDICTED: glutamate synthase 1 [NADH], chloroplastic isoform X1 [Jatropha curcas] |
| 18 |
Hb_005181_180 |
0.1472234647 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_000085_380 |
0.1482574038 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_000215_250 |
0.14934371 |
- |
- |
Acid phosphatase 1 precursor, putative [Ricinus communis] |