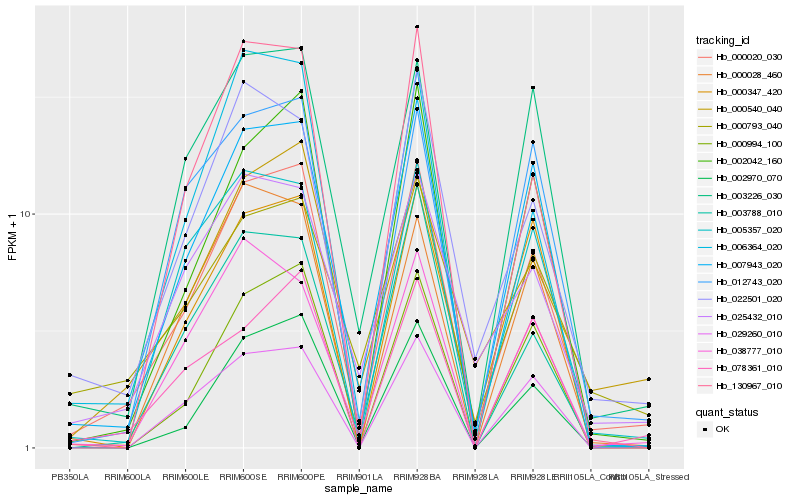
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000020_030 |
0.0 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 2 |
Hb_007943_020 |
0.1002651911 |
- |
- |
transcription factor, putative [Ricinus communis] |
| 3 |
Hb_002970_070 |
0.1150832927 |
- |
- |
multidrug/pheromone exporter protein [Hevea brasiliensis] |
| 4 |
Hb_002042_160 |
0.115474565 |
- |
- |
chloroplast-targeted copper chaperone, putative [Ricinus communis] |
| 5 |
Hb_022501_020 |
0.1164926091 |
- |
- |
AMP dependent CoA ligase, putative [Ricinus communis] |
| 6 |
Hb_003226_030 |
0.1243578744 |
- |
- |
sucrose synthase 5 [Hevea brasiliensis] |
| 7 |
Hb_005357_020 |
0.1281121801 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_000540_040 |
0.1298912858 |
- |
- |
PREDICTED: chitinase 2 [Amborella trichopoda] |
| 9 |
Hb_006364_020 |
0.1321342956 |
- |
- |
PREDICTED: leucine-rich repeat receptor-like serine/threonine-protein kinase BAM1 [Jatropha curcas] |
| 10 |
Hb_012743_020 |
0.1322855842 |
- |
- |
serine/threonine protein phosphatase, putative [Ricinus communis] |
| 11 |
Hb_130967_010 |
0.132665275 |
- |
- |
UDP-glucosyltransferase, putative [Ricinus communis] |
| 12 |
Hb_025432_010 |
0.1348812607 |
- |
- |
hypothetical protein JCGZ_21627 [Jatropha curcas] |
| 13 |
Hb_000994_100 |
0.1359388682 |
- |
- |
RALFL33, putative [Ricinus communis] |
| 14 |
Hb_000793_040 |
0.1387444088 |
- |
- |
PREDICTED: transcription factor LHW [Jatropha curcas] |
| 15 |
Hb_029260_010 |
0.1404803965 |
- |
- |
nitrate transporter, putative [Ricinus communis] |
| 16 |
Hb_038777_010 |
0.1426701345 |
transcription factor |
TF Family: G2-like |
PREDICTED: probable transcription factor KAN2 isoform X1 [Populus euphratica] |
| 17 |
Hb_003788_010 |
0.1432325731 |
- |
- |
PREDICTED: receptor-like protein kinase HSL1 [Jatropha curcas] |
| 18 |
Hb_078361_010 |
0.1436005472 |
- |
- |
PREDICTED: AT-hook motif nuclear-localized protein 1 [Jatropha curcas] |
| 19 |
Hb_000028_460 |
0.1436279725 |
- |
- |
hypothetical protein POPTR_0015s05340g [Populus trichocarpa] |
| 20 |
Hb_000347_420 |
0.1454619732 |
- |
- |
hypothetical protein JCGZ_24696 [Jatropha curcas] |