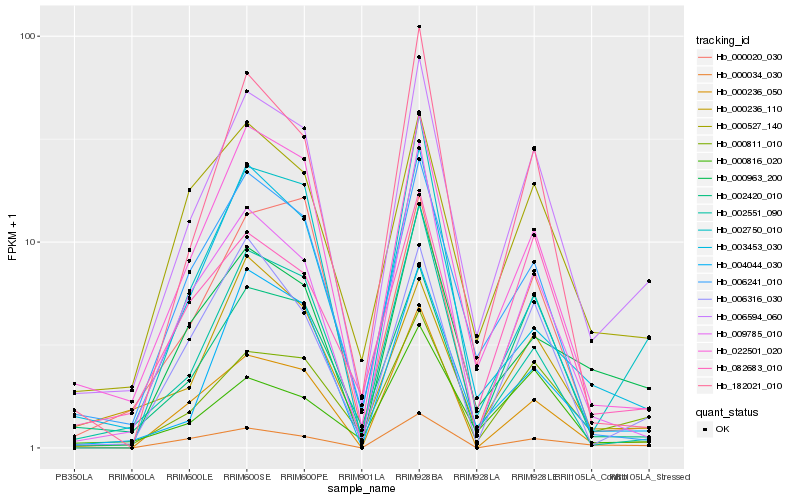
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_006594_060 |
0.0 |
transcription factor |
TF Family: HSF |
PREDICTED: heat shock factor protein HSF24 [Jatropha curcas] |
| 2 |
Hb_000811_010 |
0.1010799344 |
transcription factor |
TF Family: ERF |
PREDICTED: ethylene-responsive transcription factor ERF118 [Jatropha curcas] |
| 3 |
Hb_022501_020 |
0.115893077 |
- |
- |
AMP dependent CoA ligase, putative [Ricinus communis] |
| 4 |
Hb_000527_140 |
0.1263138519 |
transcription factor |
TF Family: GRAS |
Chitin-inducible gibberellin-responsive protein, putative [Ricinus communis] |
| 5 |
Hb_000963_200 |
0.1330272082 |
- |
- |
beta-galactosidase, putative [Ricinus communis] |
| 6 |
Hb_082683_010 |
0.1339206831 |
- |
- |
PREDICTED: uncharacterized protein LOC104898613 [Beta vulgaris subsp. vulgaris] |
| 7 |
Hb_006316_030 |
0.1400359835 |
- |
- |
aldehyde oxidase, putative [Ricinus communis] |
| 8 |
Hb_006241_010 |
0.1403726544 |
desease resistance |
Gene Name: NB-ARC |
Disease resistance protein RFL1, putative [Ricinus communis] |
| 9 |
Hb_002420_010 |
0.140941286 |
- |
- |
PREDICTED: uncharacterized protein LOC105124982 [Populus euphratica] |
| 10 |
Hb_004044_030 |
0.1426499557 |
- |
- |
PREDICTED: uncharacterized protein LOC105639335 [Jatropha curcas] |
| 11 |
Hb_182021_010 |
0.1513871509 |
- |
- |
sucrose transporter 4 [Hevea brasiliensis] |
| 12 |
Hb_000236_050 |
0.151468051 |
- |
- |
phosphate transporter [Manihot esculenta] |
| 13 |
Hb_000236_110 |
0.1514989303 |
- |
- |
PREDICTED: chaperone protein dnaJ 20, chloroplastic [Jatropha curcas] |
| 14 |
Hb_002551_090 |
0.1524415755 |
- |
- |
hypothetical protein JCGZ_17224 [Jatropha curcas] |
| 15 |
Hb_009785_010 |
0.1534615093 |
- |
- |
PREDICTED: uncharacterized protein LOC105648749 [Jatropha curcas] |
| 16 |
Hb_000034_030 |
0.1562859207 |
- |
- |
protein phosphatase 2c, putative [Ricinus communis] |
| 17 |
Hb_002750_010 |
0.1569781221 |
transcription factor |
TF Family: WRKY |
WRKY transcription factor 15 family protein [Populus trichocarpa] |
| 18 |
Hb_000020_030 |
0.158196921 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 19 |
Hb_000816_020 |
0.1600201797 |
desease resistance |
Gene Name: NB-ARC |
unnamed protein product [Coffea canephora] |
| 20 |
Hb_003453_030 |
0.1607223728 |
- |
- |
hypothetical protein JCGZ_26893 [Jatropha curcas] |