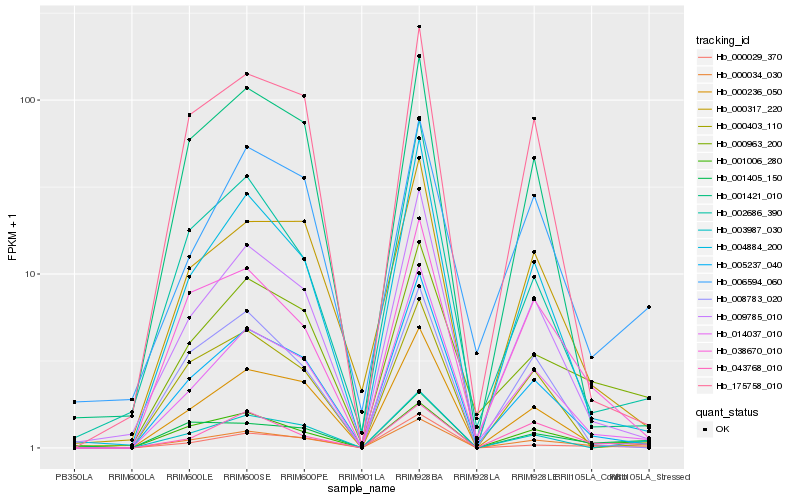
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000034_030 |
0.0 |
- |
- |
protein phosphatase 2c, putative [Ricinus communis] |
| 2 |
Hb_001006_280 |
0.0930454237 |
- |
- |
PREDICTED: glutamate decarboxylase 4 [Jatropha curcas] |
| 3 |
Hb_000963_200 |
0.1137676085 |
- |
- |
beta-galactosidase, putative [Ricinus communis] |
| 4 |
Hb_000236_050 |
0.1147940077 |
- |
- |
phosphate transporter [Manihot esculenta] |
| 5 |
Hb_038670_010 |
0.1227723257 |
- |
- |
Major allergen Pru ar, putative [Ricinus communis] |
| 6 |
Hb_005237_040 |
0.1273322522 |
- |
- |
PREDICTED: uncharacterized protein LOC105630923 [Jatropha curcas] |
| 7 |
Hb_001405_150 |
0.1333018658 |
- |
- |
PREDICTED: protein At-4/1 [Jatropha curcas] |
| 8 |
Hb_014037_010 |
0.1337118336 |
- |
- |
PREDICTED: uncharacterized protein LOC105641871 [Jatropha curcas] |
| 9 |
Hb_003987_030 |
0.135722382 |
- |
- |
PREDICTED: cytochrome P450 83B1-like [Jatropha curcas] |
| 10 |
Hb_009785_010 |
0.1402434018 |
- |
- |
PREDICTED: uncharacterized protein LOC105648749 [Jatropha curcas] |
| 11 |
Hb_043768_010 |
0.1453319206 |
- |
- |
PREDICTED: heat stress transcription factor A-4a-like [Jatropha curcas] |
| 12 |
Hb_004884_200 |
0.146135899 |
- |
- |
PREDICTED: ferritin-3, chloroplastic [Jatropha curcas] |
| 13 |
Hb_000317_220 |
0.1467200677 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_000029_370 |
0.1536155281 |
- |
- |
multidrug resistance pump, putative [Ricinus communis] |
| 15 |
Hb_006594_060 |
0.1562859207 |
transcription factor |
TF Family: HSF |
PREDICTED: heat shock factor protein HSF24 [Jatropha curcas] |
| 16 |
Hb_000403_110 |
0.1568950439 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 51 [Jatropha curcas] |
| 17 |
Hb_175758_010 |
0.1605501182 |
- |
- |
PREDICTED: aspartic proteinase A1-like isoform X2 [Jatropha curcas] |
| 18 |
Hb_008783_020 |
0.1644034666 |
- |
- |
hypothetical protein JCGZ_21284 [Jatropha curcas] |
| 19 |
Hb_002686_390 |
0.1653399537 |
- |
- |
PREDICTED: uncharacterized protein LOC105647789 [Jatropha curcas] |
| 20 |
Hb_001421_010 |
0.1658579716 |
transcription factor |
TF Family: ARR-B |
PREDICTED: two-component response regulator-like APRR2 isoform X1 [Jatropha curcas] |