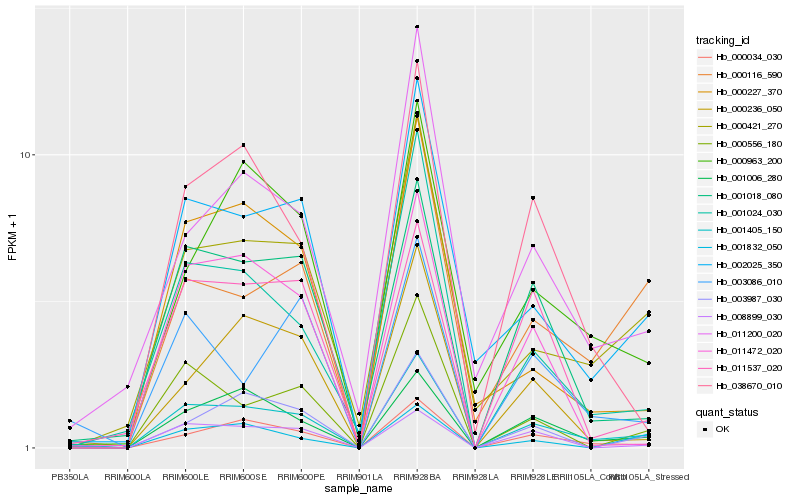
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001405_150 |
0.0 |
- |
- |
PREDICTED: protein At-4/1 [Jatropha curcas] |
| 2 |
Hb_000034_030 |
0.1333018658 |
- |
- |
protein phosphatase 2c, putative [Ricinus communis] |
| 3 |
Hb_001006_280 |
0.1442795426 |
- |
- |
PREDICTED: glutamate decarboxylase 4 [Jatropha curcas] |
| 4 |
Hb_002025_350 |
0.1477267554 |
- |
- |
PREDICTED: glutamate decarboxylase 4 [Jatropha curcas] |
| 5 |
Hb_001024_030 |
0.155444834 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RGLG2-like isoform X1 [Jatropha curcas] |
| 6 |
Hb_001018_080 |
0.1577599385 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_000421_270 |
0.1642377355 |
transcription factor |
TF Family: LOB |
LOB domain-containing protein 1 [Theobroma cacao] |
| 8 |
Hb_000116_590 |
0.1703678519 |
- |
- |
PREDICTED: splicing factor, suppressor of white-apricot homolog [Malus domestica] |
| 9 |
Hb_011200_020 |
0.1709152207 |
- |
- |
aldo-keto reductase, putative [Ricinus communis] |
| 10 |
Hb_038670_010 |
0.1724085762 |
- |
- |
Major allergen Pru ar, putative [Ricinus communis] |
| 11 |
Hb_003086_010 |
0.1727488231 |
- |
- |
hypothetical protein JCGZ_06093 [Jatropha curcas] |
| 12 |
Hb_000556_180 |
0.1772845451 |
- |
- |
hypothetical protein POPTR_0014s06690g [Populus trichocarpa] |
| 13 |
Hb_003987_030 |
0.1775448021 |
- |
- |
PREDICTED: cytochrome P450 83B1-like [Jatropha curcas] |
| 14 |
Hb_000963_200 |
0.1779344285 |
- |
- |
beta-galactosidase, putative [Ricinus communis] |
| 15 |
Hb_000227_370 |
0.1810312979 |
- |
- |
sugar transporter, putative [Ricinus communis] |
| 16 |
Hb_000236_050 |
0.1811749023 |
- |
- |
phosphate transporter [Manihot esculenta] |
| 17 |
Hb_008899_030 |
0.1829158643 |
- |
- |
Lipase class 3-related protein, putative [Theobroma cacao] |
| 18 |
Hb_001832_050 |
0.1839984929 |
- |
- |
PREDICTED: cytokinin dehydrogenase 3 [Jatropha curcas] |
| 19 |
Hb_011472_020 |
0.1898629706 |
- |
- |
PREDICTED: mucin-17-like [Jatropha curcas] |
| 20 |
Hb_011537_020 |
0.190223211 |
- |
- |
PREDICTED: uncharacterized serine-rich protein C215.13-like [Jatropha curcas] |