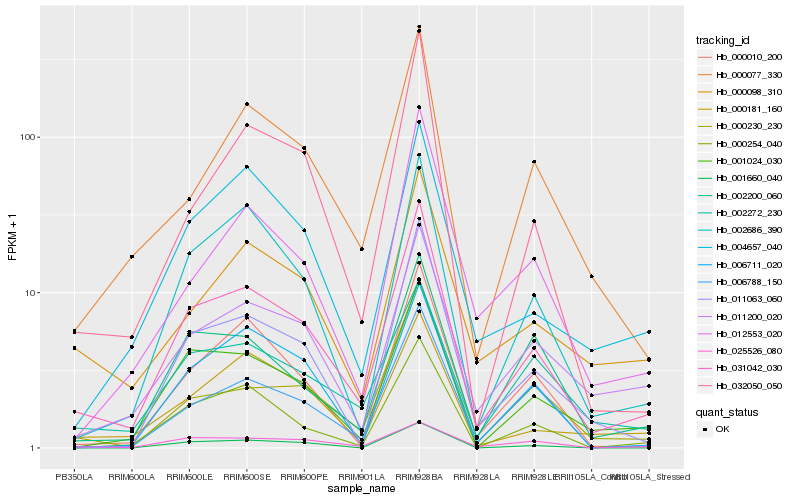
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_031042_030 |
0.0 |
- |
- |
PREDICTED: WAT1-related protein At5g47470-like isoform X1 [Jatropha curcas] |
| 2 |
Hb_001024_030 |
0.1225716081 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RGLG2-like isoform X1 [Jatropha curcas] |
| 3 |
Hb_002686_390 |
0.129923254 |
- |
- |
PREDICTED: uncharacterized protein LOC105647789 [Jatropha curcas] |
| 4 |
Hb_011063_060 |
0.1329544784 |
- |
- |
PREDICTED: endochitinase-like [Jatropha curcas] |
| 5 |
Hb_000230_230 |
0.1351798193 |
- |
- |
hydrolase, putative [Ricinus communis] |
| 6 |
Hb_011200_020 |
0.1367894092 |
- |
- |
aldo-keto reductase, putative [Ricinus communis] |
| 7 |
Hb_000254_040 |
0.1395184021 |
- |
- |
PREDICTED: uncharacterized protein LOC105634992 [Jatropha curcas] |
| 8 |
Hb_000077_330 |
0.1402331189 |
- |
- |
PREDICTED: bifunctional epoxide hydrolase 2-like [Jatropha curcas] |
| 9 |
Hb_032050_050 |
0.140318861 |
- |
- |
PREDICTED: peroxidase 12-like [Jatropha curcas] |
| 10 |
Hb_000010_200 |
0.1434227153 |
- |
- |
BRASSINOSTEROID INSENSITIVE 1-associated receptor kinase 1 precursor, putative [Ricinus communis] |
| 11 |
Hb_006788_150 |
0.1460049597 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_000098_310 |
0.1500382916 |
- |
- |
RecName: Full=Anthocyanidin 3-O-glucosyltransferase 1; AltName: Full=Flavonol 3-O-glucosyltransferase 1; AltName: Full=UDP-glucose flavonoid 3-O-glucosyltransferase 1 [Manihot esculenta] |
| 13 |
Hb_002272_230 |
0.1505902107 |
- |
- |
sodium/hydrogen exchanger, putative [Ricinus communis] |
| 14 |
Hb_002200_060 |
0.1516791449 |
- |
- |
PREDICTED: pleckstrin homology domain-containing family A member 8 [Jatropha curcas] |
| 15 |
Hb_000181_160 |
0.153596985 |
- |
- |
PREDICTED: uncharacterized protein LOC105637575 isoform X2 [Jatropha curcas] |
| 16 |
Hb_012553_020 |
0.1544492511 |
- |
- |
hypothetical protein JCGZ_09282 [Jatropha curcas] |
| 17 |
Hb_025526_080 |
0.1553001451 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase At4g27290 [Vitis vinifera] |
| 18 |
Hb_004657_040 |
0.156142062 |
transcription factor |
TF Family: WRKY |
WRKY transcription factor 15 family protein [Populus trichocarpa] |
| 19 |
Hb_001660_040 |
0.1592446053 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At4g08850 [Jatropha curcas] |
| 20 |
Hb_006711_020 |
0.159916429 |
- |
- |
hypothetical protein JCGZ_07258 [Jatropha curcas] |