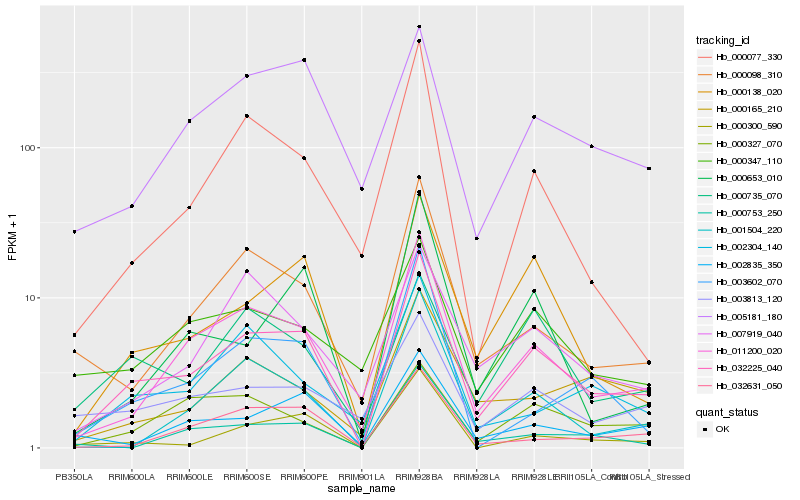
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_032225_040 |
0.0 |
- |
- |
- |
| 2 |
Hb_011200_020 |
0.1283162328 |
- |
- |
aldo-keto reductase, putative [Ricinus communis] |
| 3 |
Hb_000138_020 |
0.1570922723 |
- |
- |
PREDICTED: peroxidase A2-like [Jatropha curcas] |
| 4 |
Hb_000098_310 |
0.1785240135 |
- |
- |
RecName: Full=Anthocyanidin 3-O-glucosyltransferase 1; AltName: Full=Flavonol 3-O-glucosyltransferase 1; AltName: Full=UDP-glucose flavonoid 3-O-glucosyltransferase 1 [Manihot esculenta] |
| 5 |
Hb_000300_590 |
0.1812984409 |
transcription factor |
TF Family: ERF |
PREDICTED: ethylene-responsive transcription factor RAP2-11-like [Jatropha curcas] |
| 6 |
Hb_002835_350 |
0.1862027115 |
- |
- |
PREDICTED: uncharacterized protein LOC105640319 [Jatropha curcas] |
| 7 |
Hb_032631_050 |
0.1865164099 |
- |
- |
PREDICTED: probable inactive leucine-rich repeat receptor-like protein kinase At1g66830 [Jatropha curcas] |
| 8 |
Hb_001504_220 |
0.1900515396 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |
| 9 |
Hb_003813_120 |
0.1921800333 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 10 |
Hb_000077_330 |
0.1971811995 |
- |
- |
PREDICTED: bifunctional epoxide hydrolase 2-like [Jatropha curcas] |
| 11 |
Hb_000735_070 |
0.19771373 |
transcription factor |
TF Family: bHLH |
basic helix-loop-helix family protein [Populus trichocarpa] |
| 12 |
Hb_000653_010 |
0.1979772248 |
- |
- |
hypothetical protein CICLE_v10033238mg [Citrus clementina] |
| 13 |
Hb_003602_070 |
0.1992291265 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor bHLH93-like [Jatropha curcas] |
| 14 |
Hb_005181_180 |
0.1999166128 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_000753_250 |
0.2005302783 |
- |
- |
PREDICTED: uncharacterized protein LOC105640661 [Jatropha curcas] |
| 16 |
Hb_000165_210 |
0.2013415157 |
- |
- |
PREDICTED: trihelix transcription factor ASIL2 isoform X2 [Jatropha curcas] |
| 17 |
Hb_000347_110 |
0.2013828726 |
- |
- |
BnaA05g25330D [Brassica napus] |
| 18 |
Hb_000327_070 |
0.2021223635 |
transcription factor |
TF Family: ERF |
PREDICTED: AP2-like ethylene-responsive transcription factor TOE3 isoform X2 [Jatropha curcas] |
| 19 |
Hb_002304_140 |
0.2041114055 |
- |
- |
ring finger protein, putative [Ricinus communis] |
| 20 |
Hb_007919_040 |
0.20474092 |
- |
- |
PREDICTED: protein LYK5 [Jatropha curcas] |