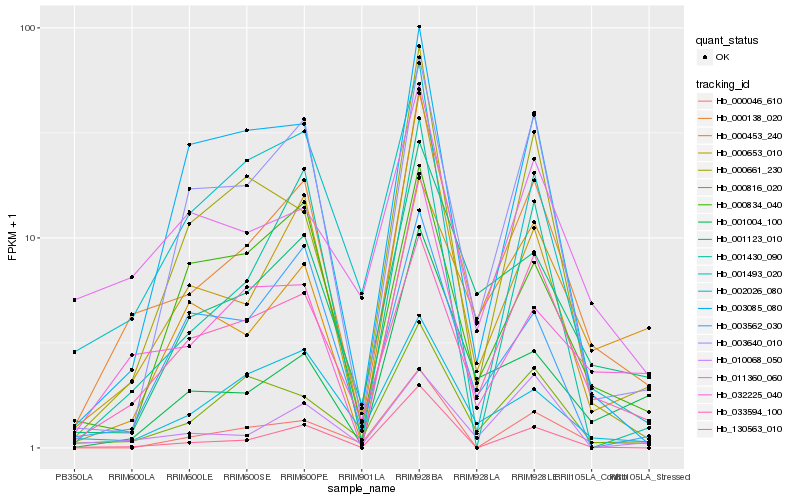
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000138_020 |
0.0 |
- |
- |
PREDICTED: peroxidase A2-like [Jatropha curcas] |
| 2 |
Hb_000653_010 |
0.1436857499 |
- |
- |
hypothetical protein CICLE_v10033238mg [Citrus clementina] |
| 3 |
Hb_001123_010 |
0.1533258885 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_033594_100 |
0.1541413102 |
transcription factor |
TF Family: GNAT |
PREDICTED: probable amino-acid acetyltransferase NAGS1, chloroplastic isoform X1 [Jatropha curcas] |
| 5 |
Hb_032225_040 |
0.1570922723 |
- |
- |
- |
| 6 |
Hb_000816_020 |
0.1696799994 |
desease resistance |
Gene Name: NB-ARC |
unnamed protein product [Coffea canephora] |
| 7 |
Hb_003640_010 |
0.1716310102 |
- |
- |
interferon-induced GTP-binding protein mx, putative [Ricinus communis] |
| 8 |
Hb_002026_080 |
0.1824573944 |
desease resistance |
Gene Name: TIR_2 |
nucleoside-triphosphatase, putative [Ricinus communis] |
| 9 |
Hb_130563_010 |
0.1886733648 |
- |
- |
PREDICTED: ABC transporter C family member 3-like isoform X1 [Jatropha curcas] |
| 10 |
Hb_001430_090 |
0.1925143491 |
- |
- |
hypothetical protein JCGZ_05131 [Jatropha curcas] |
| 11 |
Hb_011360_060 |
0.1943250476 |
- |
- |
glycogen phosphorylase, putative [Ricinus communis] |
| 12 |
Hb_000834_040 |
0.1983298084 |
- |
- |
PREDICTED: ras-related protein Rab7 [Cucumis melo] |
| 13 |
Hb_000661_230 |
0.199344321 |
- |
- |
PREDICTED: zinc finger CCCH domain-containing protein 22-like isoform X2 [Jatropha curcas] |
| 14 |
Hb_000453_240 |
0.2001374772 |
rubber biosynthesis |
Gene Name: 4-(cytidine 5'-diphospho)-2-C-methyl-D-erythritol kinase |
4-(cytidine 5'-diphospho)-2-C-methyl-D-erythritol kinase [Hevea brasiliensis] |
| 15 |
Hb_003085_080 |
0.2006104921 |
- |
- |
nucleic acid binding protein, putative [Ricinus communis] |
| 16 |
Hb_001004_100 |
0.2019773794 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 17 |
Hb_000046_610 |
0.2025979807 |
- |
- |
PREDICTED: putative cullin-like protein 2 isoform X3 [Populus euphratica] |
| 18 |
Hb_003562_030 |
0.2034261449 |
- |
- |
- |
| 19 |
Hb_001493_020 |
0.2046252654 |
- |
- |
leucine-rich repeat receptor-like protein kinase [Manihot esculenta] |
| 20 |
Hb_010068_050 |
0.2048763307 |
- |
- |
PREDICTED: uncharacterized protein LOC105632963 [Jatropha curcas] |