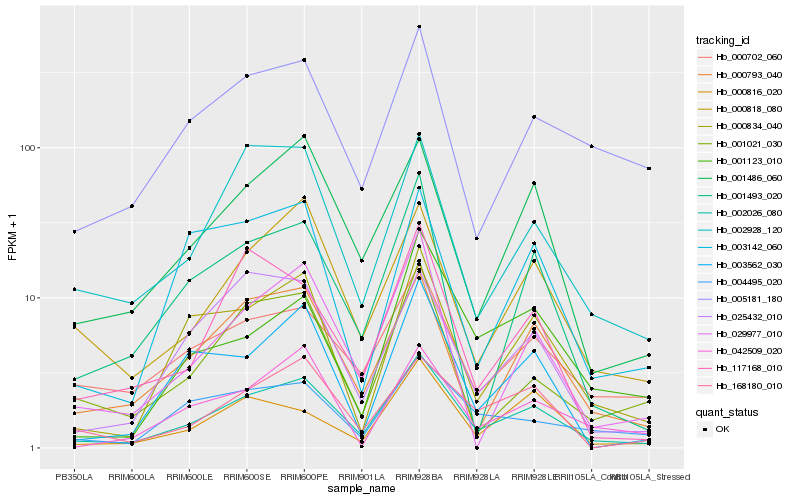
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002026_080 |
0.0 |
desease resistance |
Gene Name: TIR_2 |
nucleoside-triphosphatase, putative [Ricinus communis] |
| 2 |
Hb_000793_040 |
0.1306680374 |
- |
- |
PREDICTED: transcription factor LHW [Jatropha curcas] |
| 3 |
Hb_001486_060 |
0.1326508069 |
- |
- |
PREDICTED: uncharacterized protein LOC105632622 [Jatropha curcas] |
| 4 |
Hb_000834_040 |
0.1376023224 |
- |
- |
PREDICTED: ras-related protein Rab7 [Cucumis melo] |
| 5 |
Hb_000818_080 |
0.1388151299 |
- |
- |
PREDICTED: monosaccharide-sensing protein 2 [Jatropha curcas] |
| 6 |
Hb_000702_060 |
0.1461847448 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_001493_020 |
0.1485732954 |
- |
- |
leucine-rich repeat receptor-like protein kinase [Manihot esculenta] |
| 8 |
Hb_002928_120 |
0.1486764624 |
- |
- |
1,4-alpha-glucan branching enzyme [Manihot esculenta] |
| 9 |
Hb_029977_010 |
0.1530629585 |
- |
- |
catalytic, putative [Ricinus communis] |
| 10 |
Hb_168180_010 |
0.1542735219 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_003562_030 |
0.1592054556 |
- |
- |
- |
| 12 |
Hb_001021_030 |
0.1612999383 |
- |
- |
chloroplast-targeted copper chaperone, putative [Ricinus communis] |
| 13 |
Hb_005181_180 |
0.1634307234 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_025432_010 |
0.1639091894 |
- |
- |
hypothetical protein JCGZ_21627 [Jatropha curcas] |
| 15 |
Hb_042509_020 |
0.1655632752 |
- |
- |
hypothetical protein JCGZ_19555 [Jatropha curcas] |
| 16 |
Hb_004495_020 |
0.1662383689 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance protein RGA3 [Jatropha curcas] |
| 17 |
Hb_117168_010 |
0.171804351 |
- |
- |
PREDICTED: putative disease resistance protein RGA1 [Jatropha curcas] |
| 18 |
Hb_001123_010 |
0.1718595046 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_000816_020 |
0.1726388143 |
desease resistance |
Gene Name: NB-ARC |
unnamed protein product [Coffea canephora] |
| 20 |
Hb_003142_060 |
0.1735511201 |
- |
- |
PREDICTED: glutamate synthase 1 [NADH], chloroplastic isoform X1 [Jatropha curcas] |