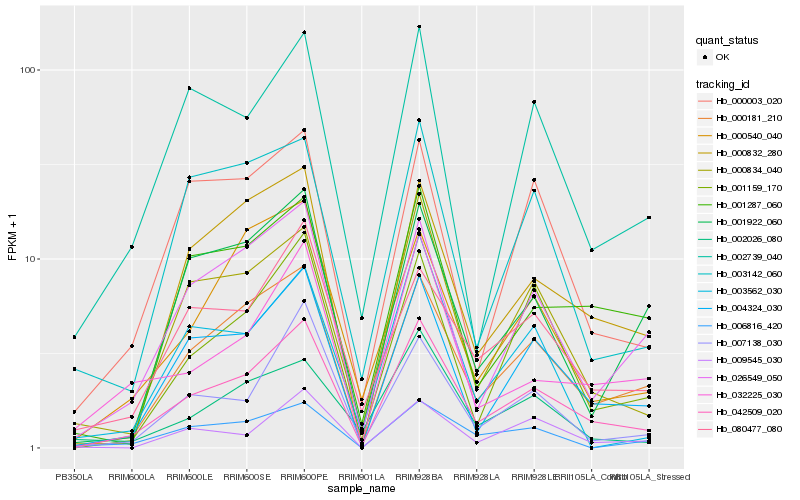
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_042509_020 |
0.0 |
- |
- |
hypothetical protein JCGZ_19555 [Jatropha curcas] |
| 2 |
Hb_000181_210 |
0.1027287571 |
- |
- |
Phytosulfokine receptor precursor, putative [Ricinus communis] |
| 3 |
Hb_004324_030 |
0.1028647763 |
- |
- |
PREDICTED: probable protein phosphatase 2C 8 [Jatropha curcas] |
| 4 |
Hb_000834_040 |
0.1334173932 |
- |
- |
PREDICTED: ras-related protein Rab7 [Cucumis melo] |
| 5 |
Hb_000832_280 |
0.1400935186 |
- |
- |
PREDICTED: aquaporin TIP1-3 [Jatropha curcas] |
| 6 |
Hb_026549_050 |
0.1410559859 |
- |
- |
PREDICTED: uncharacterized protein LOC105636246 isoform X2 [Jatropha curcas] |
| 7 |
Hb_001159_170 |
0.1476303486 |
- |
- |
PREDICTED: AT-hook motif nuclear-localized protein 1 [Jatropha curcas] |
| 8 |
Hb_007138_030 |
0.1501165797 |
- |
- |
PREDICTED: putative disease resistance protein At4g11170 [Jatropha curcas] |
| 9 |
Hb_080477_080 |
0.1503173575 |
- |
- |
PREDICTED: protein NUCLEAR FUSION DEFECTIVE 4 isoform X1 [Gossypium raimondii] |
| 10 |
Hb_002739_040 |
0.1518250061 |
- |
- |
PREDICTED: isocitrate dehydrogenase [NADP] [Jatropha curcas] |
| 11 |
Hb_032225_030 |
0.1526128881 |
- |
- |
PREDICTED: leucine-rich repeat extensin-like protein 2 [Jatropha curcas] |
| 12 |
Hb_001287_060 |
0.1571056532 |
transcription factor |
TF Family: Orphans |
Salt-tolerance protein, putative [Ricinus communis] |
| 13 |
Hb_000540_040 |
0.1603490559 |
- |
- |
PREDICTED: chitinase 2 [Amborella trichopoda] |
| 14 |
Hb_000003_020 |
0.1611737841 |
- |
- |
PREDICTED: uncharacterized protein LOC105631161 [Jatropha curcas] |
| 15 |
Hb_009545_030 |
0.1637779399 |
- |
- |
PREDICTED: folate-biopterin transporter 1, chloroplastic [Jatropha curcas] |
| 16 |
Hb_002026_080 |
0.1655632752 |
desease resistance |
Gene Name: TIR_2 |
nucleoside-triphosphatase, putative [Ricinus communis] |
| 17 |
Hb_001922_060 |
0.1692395932 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 18 |
Hb_003142_060 |
0.170741816 |
- |
- |
PREDICTED: glutamate synthase 1 [NADH], chloroplastic isoform X1 [Jatropha curcas] |
| 19 |
Hb_006816_420 |
0.1720151409 |
- |
- |
hypothetical protein JCGZ_23261 [Jatropha curcas] |
| 20 |
Hb_003562_030 |
0.1759036969 |
- |
- |
- |