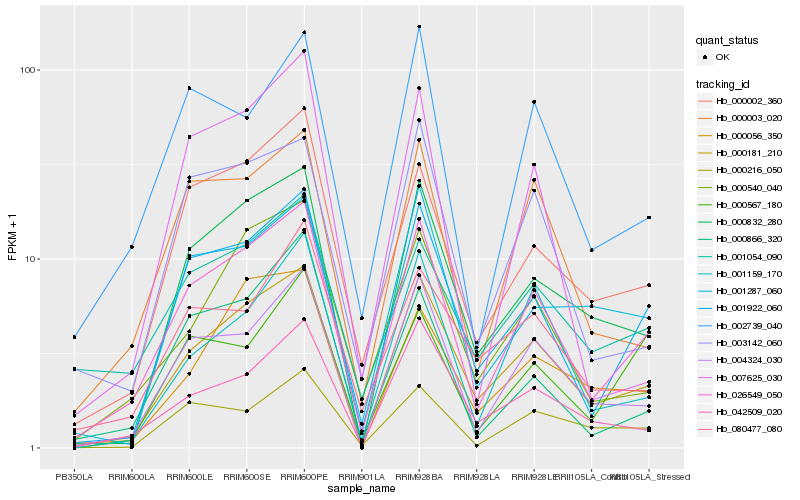
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_026549_050 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105636246 isoform X2 [Jatropha curcas] |
| 2 |
Hb_000181_210 |
0.0880127687 |
- |
- |
Phytosulfokine receptor precursor, putative [Ricinus communis] |
| 3 |
Hb_001922_060 |
0.0918963157 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 4 |
Hb_004324_030 |
0.1055529885 |
- |
- |
PREDICTED: probable protein phosphatase 2C 8 [Jatropha curcas] |
| 5 |
Hb_000002_360 |
0.1231827163 |
- |
- |
hypothetical protein POPTR_0008s16470g [Populus trichocarpa] |
| 6 |
Hb_000003_020 |
0.1281492604 |
- |
- |
PREDICTED: uncharacterized protein LOC105631161 [Jatropha curcas] |
| 7 |
Hb_000832_280 |
0.1313214666 |
- |
- |
PREDICTED: aquaporin TIP1-3 [Jatropha curcas] |
| 8 |
Hb_002739_040 |
0.1339333923 |
- |
- |
PREDICTED: isocitrate dehydrogenase [NADP] [Jatropha curcas] |
| 9 |
Hb_000540_040 |
0.1382869084 |
- |
- |
PREDICTED: chitinase 2 [Amborella trichopoda] |
| 10 |
Hb_042509_020 |
0.1410559859 |
- |
- |
hypothetical protein JCGZ_19555 [Jatropha curcas] |
| 11 |
Hb_003142_060 |
0.1422268058 |
- |
- |
PREDICTED: glutamate synthase 1 [NADH], chloroplastic isoform X1 [Jatropha curcas] |
| 12 |
Hb_000216_050 |
0.1491326728 |
- |
- |
PREDICTED: indole-3-acetaldehyde oxidase-like [Jatropha curcas] |
| 13 |
Hb_001054_090 |
0.1494902583 |
- |
- |
Potassium channel SKOR, putative [Ricinus communis] |
| 14 |
Hb_000866_320 |
0.1554465065 |
- |
- |
receptor protein kinase, putative [Ricinus communis] |
| 15 |
Hb_001159_170 |
0.1568309904 |
- |
- |
PREDICTED: AT-hook motif nuclear-localized protein 1 [Jatropha curcas] |
| 16 |
Hb_001287_060 |
0.1585128489 |
transcription factor |
TF Family: Orphans |
Salt-tolerance protein, putative [Ricinus communis] |
| 17 |
Hb_007625_030 |
0.1615390767 |
- |
- |
unknown [Medicago truncatula] |
| 18 |
Hb_000056_350 |
0.164104917 |
- |
- |
PREDICTED: uncharacterized protein LOC105630437 [Jatropha curcas] |
| 19 |
Hb_000567_180 |
0.1654547622 |
transcription factor |
TF Family: C2C2-Dof |
PREDICTED: dof zinc finger protein DOF1.5 [Jatropha curcas] |
| 20 |
Hb_080477_080 |
0.1656562185 |
- |
- |
PREDICTED: protein NUCLEAR FUSION DEFECTIVE 4 isoform X1 [Gossypium raimondii] |