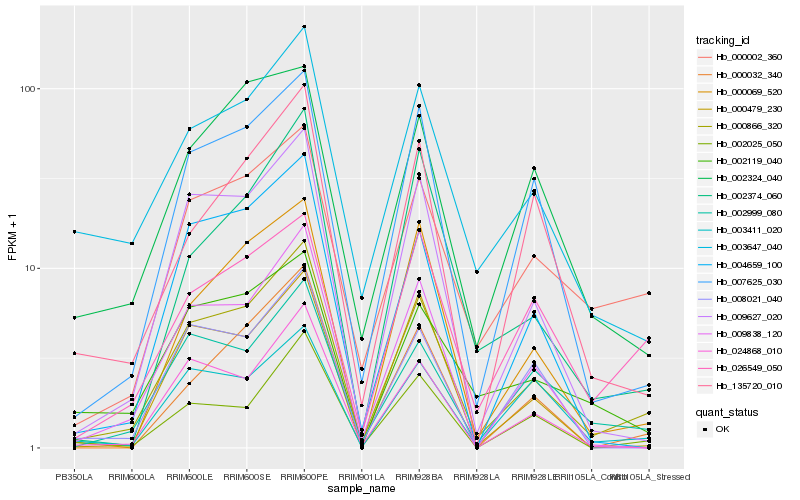
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000866_320 |
0.0 |
- |
- |
receptor protein kinase, putative [Ricinus communis] |
| 2 |
Hb_009627_020 |
0.1120081515 |
- |
- |
PREDICTED: probable pectinesterase/pectinesterase inhibitor 34 [Jatropha curcas] |
| 3 |
Hb_007625_030 |
0.1180506419 |
- |
- |
unknown [Medicago truncatula] |
| 4 |
Hb_009838_120 |
0.1184432742 |
transcription factor |
TF Family: AUX/IAA |
PREDICTED: auxin-responsive protein IAA13 isoform X1 [Jatropha curcas] |
| 5 |
Hb_002999_080 |
0.1268844465 |
transcription factor |
TF Family: G2-like |
PREDICTED: accessory gland protein Acp36DE [Jatropha curcas] |
| 6 |
Hb_000002_360 |
0.1296081271 |
- |
- |
hypothetical protein POPTR_0008s16470g [Populus trichocarpa] |
| 7 |
Hb_004659_100 |
0.1357025955 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_003647_040 |
0.1358674869 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_024868_010 |
0.1391458186 |
- |
- |
protein kinase, putative [Ricinus communis] |
| 10 |
Hb_002025_050 |
0.1394530247 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_000069_520 |
0.1421184423 |
- |
- |
PREDICTED: transcription factor TRY-like [Jatropha curcas] |
| 12 |
Hb_000032_340 |
0.1465560258 |
- |
- |
PREDICTED: uncharacterized protein LOC105645669 [Jatropha curcas] |
| 13 |
Hb_000479_230 |
0.1524888948 |
- |
- |
hypothetical protein JCGZ_19903 [Jatropha curcas] |
| 14 |
Hb_002119_040 |
0.1546625072 |
- |
- |
receptor protein kinase, putative [Ricinus communis] |
| 15 |
Hb_002374_060 |
0.1551274927 |
- |
- |
amino acid transporter, putative [Ricinus communis] |
| 16 |
Hb_026549_050 |
0.1554465065 |
- |
- |
PREDICTED: uncharacterized protein LOC105636246 isoform X2 [Jatropha curcas] |
| 17 |
Hb_008021_040 |
0.155820711 |
- |
- |
PREDICTED: AP2-like ethylene-responsive transcription factor ANT [Jatropha curcas] |
| 18 |
Hb_003411_020 |
0.1568733001 |
- |
- |
alpha-glucosidase, putative [Ricinus communis] |
| 19 |
Hb_002324_040 |
0.1570977627 |
- |
- |
PREDICTED: subtilisin-like protease SBT1.6 [Jatropha curcas] |
| 20 |
Hb_135720_010 |
0.1574396351 |
- |
- |
hypothetical protein JCGZ_17857 [Jatropha curcas] |