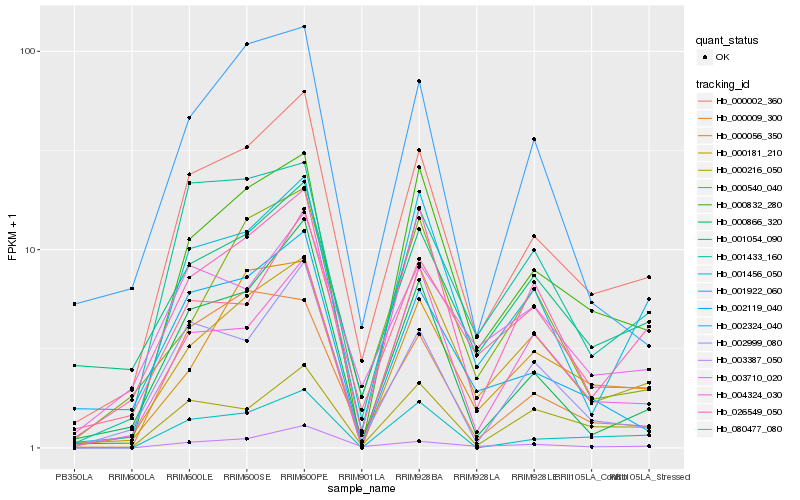
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000002_360 |
0.0 |
- |
- |
hypothetical protein POPTR_0008s16470g [Populus trichocarpa] |
| 2 |
Hb_026549_050 |
0.1231827163 |
- |
- |
PREDICTED: uncharacterized protein LOC105636246 isoform X2 [Jatropha curcas] |
| 3 |
Hb_000832_280 |
0.1275190905 |
- |
- |
PREDICTED: aquaporin TIP1-3 [Jatropha curcas] |
| 4 |
Hb_000866_320 |
0.1296081271 |
- |
- |
receptor protein kinase, putative [Ricinus communis] |
| 5 |
Hb_001433_160 |
0.1314757727 |
- |
- |
phosphatase 2C family protein [Populus trichocarpa] |
| 6 |
Hb_003387_050 |
0.1318412914 |
- |
- |
PREDICTED: uncharacterized protein LOC105639324 [Jatropha curcas] |
| 7 |
Hb_001922_060 |
0.1356914445 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 8 |
Hb_002999_080 |
0.1363350315 |
transcription factor |
TF Family: G2-like |
PREDICTED: accessory gland protein Acp36DE [Jatropha curcas] |
| 9 |
Hb_001054_090 |
0.1365729071 |
- |
- |
Potassium channel SKOR, putative [Ricinus communis] |
| 10 |
Hb_000181_210 |
0.1402709548 |
- |
- |
Phytosulfokine receptor precursor, putative [Ricinus communis] |
| 11 |
Hb_080477_080 |
0.1448992563 |
- |
- |
PREDICTED: protein NUCLEAR FUSION DEFECTIVE 4 isoform X1 [Gossypium raimondii] |
| 12 |
Hb_000216_050 |
0.1466846414 |
- |
- |
PREDICTED: indole-3-acetaldehyde oxidase-like [Jatropha curcas] |
| 13 |
Hb_003710_020 |
0.1484770239 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At1g53440 isoform X1 [Jatropha curcas] |
| 14 |
Hb_002119_040 |
0.150017605 |
- |
- |
receptor protein kinase, putative [Ricinus communis] |
| 15 |
Hb_002324_040 |
0.1567587937 |
- |
- |
PREDICTED: subtilisin-like protease SBT1.6 [Jatropha curcas] |
| 16 |
Hb_000056_350 |
0.1569572639 |
- |
- |
PREDICTED: uncharacterized protein LOC105630437 [Jatropha curcas] |
| 17 |
Hb_004324_030 |
0.1590132821 |
- |
- |
PREDICTED: probable protein phosphatase 2C 8 [Jatropha curcas] |
| 18 |
Hb_000540_040 |
0.1592094036 |
- |
- |
PREDICTED: chitinase 2 [Amborella trichopoda] |
| 19 |
Hb_001456_050 |
0.1593644182 |
transcription factor |
TF Family: G2-like |
transcription factor, putative [Ricinus communis] |
| 20 |
Hb_000009_300 |
0.1594367801 |
- |
- |
hypothetical protein CICLE_v10024731mg [Citrus clementina] |