| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
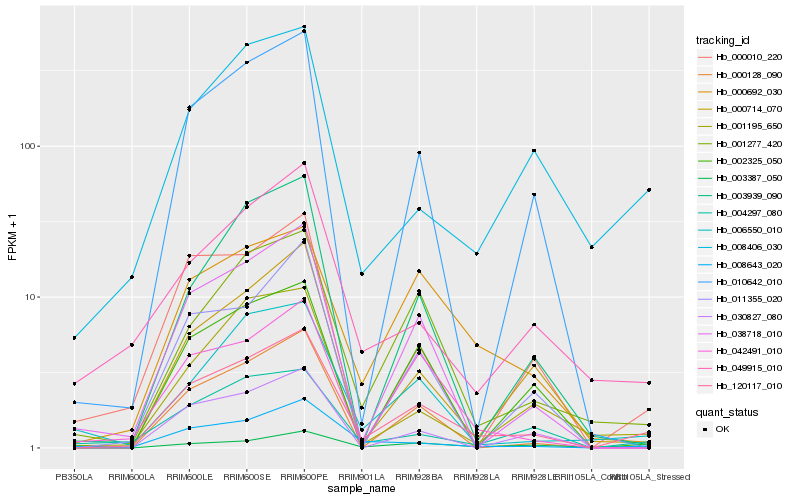
| 1 |
Hb_120117_010 |
0.0 |
- |
- |
Disease resistance protein RFL1, putative [Ricinus communis] |
| 2 |
Hb_008643_020 |
0.1574608835 |
- |
- |
Cf-4/9 disease resistance-like family protein [Populus trichocarpa] |
| 3 |
Hb_001277_420 |
0.1630929089 |
- |
- |
PREDICTED: protein lin-12-like isoform X2 [Populus euphratica] |
| 4 |
Hb_038718_010 |
0.1668587216 |
- |
- |
Disease resistance protein RFL1, putative [Ricinus communis] |
| 5 |
Hb_010642_010 |
0.1701450087 |
- |
- |
PREDICTED: aquaporin TIP2-1 [Jatropha curcas] |
| 6 |
Hb_004297_080 |
0.1736034775 |
- |
- |
PREDICTED: UPF0481 protein At3g47200-like [Jatropha curcas] |
| 7 |
Hb_000128_090 |
0.176010824 |
- |
- |
Reticuline oxidase precursor, putative [Ricinus communis] |
| 8 |
Hb_000714_070 |
0.1773079075 |
transcription factor |
TF Family: LOB |
hypothetical protein JCGZ_14754 [Jatropha curcas] |
| 9 |
Hb_042491_010 |
0.1789043651 |
- |
- |
JHL06P13.14 [Jatropha curcas] |
| 10 |
Hb_006550_010 |
0.1789575027 |
- |
- |
PREDICTED: vacuolar amino acid transporter 1 [Eucalyptus grandis] |
| 11 |
Hb_000692_030 |
0.1791707302 |
- |
- |
nucleoredoxin, putative [Ricinus communis] |
| 12 |
Hb_003387_050 |
0.1825943961 |
- |
- |
PREDICTED: uncharacterized protein LOC105639324 [Jatropha curcas] |
| 13 |
Hb_049915_010 |
0.1841426494 |
- |
- |
PREDICTED: uncharacterized serine-rich protein C215.13 [Jatropha curcas] |
| 14 |
Hb_000010_220 |
0.1848059832 |
- |
- |
PREDICTED: uncharacterized protein LOC105640277 [Jatropha curcas] |
| 15 |
Hb_008406_030 |
0.1859845398 |
transcription factor |
TF Family: ERF |
AP2/ERF super family protein [Hevea brasiliensis] |
| 16 |
Hb_003939_090 |
0.1864307866 |
transcription factor |
TF Family: NAC |
transcription factor, putative [Ricinus communis] |
| 17 |
Hb_001195_650 |
0.1877796933 |
- |
- |
PREDICTED: calcium uniporter protein 4, mitochondrial [Jatropha curcas] |
| 18 |
Hb_030827_080 |
0.188597833 |
- |
- |
PREDICTED: uncharacterized protein LOC105648491 [Jatropha curcas] |
| 19 |
Hb_002325_050 |
0.1892143438 |
transcription factor |
TF Family: MYB |
MYB transcriptional factor [Populus nigra] |
| 20 |
Hb_011355_020 |
0.1893849413 |
- |
- |
extensin, proline-rich protein, putative [Ricinus communis] |