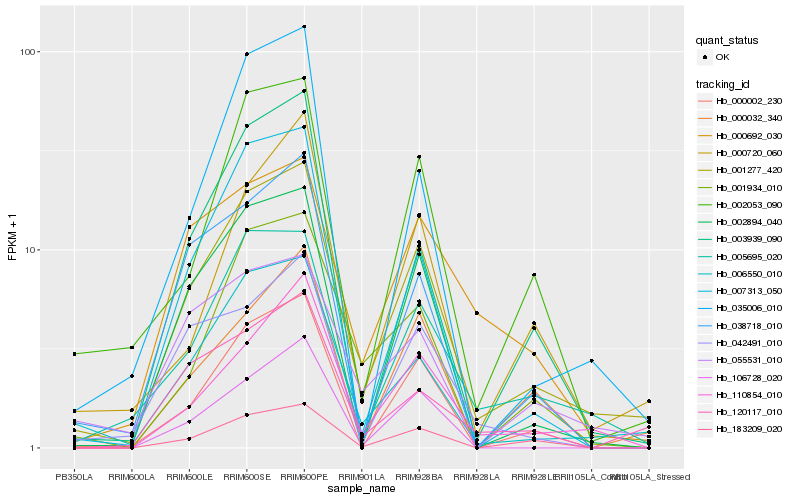
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001277_420 |
0.0 |
- |
- |
PREDICTED: protein lin-12-like isoform X2 [Populus euphratica] |
| 2 |
Hb_006550_010 |
0.1132294596 |
- |
- |
PREDICTED: vacuolar amino acid transporter 1 [Eucalyptus grandis] |
| 3 |
Hb_005695_020 |
0.1300165481 |
- |
- |
PREDICTED: low affinity sulfate transporter 3 [Jatropha curcas] |
| 4 |
Hb_002894_040 |
0.1396236664 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_000002_230 |
0.1422768365 |
- |
- |
RING/U-box superfamily protein [Theobroma cacao] |
| 6 |
Hb_183209_020 |
0.1485605398 |
- |
- |
PREDICTED: probable leucine-rich repeat receptor-like protein kinase At5g63930 isoform X2 [Jatropha curcas] |
| 7 |
Hb_002053_090 |
0.1518900629 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_007313_050 |
0.155735271 |
- |
- |
Histidine-containing phosphotransfer protein, putative [Ricinus communis] |
| 9 |
Hb_110854_010 |
0.1562729603 |
- |
- |
- |
| 10 |
Hb_001934_010 |
0.1587843294 |
- |
- |
o-methyltransferase, putative [Ricinus communis] |
| 11 |
Hb_055531_010 |
0.1595481167 |
- |
- |
PREDICTED: probable disease resistance protein At4g27220 isoform X2 [Pyrus x bretschneideri] |
| 12 |
Hb_003939_090 |
0.1610466108 |
transcription factor |
TF Family: NAC |
transcription factor, putative [Ricinus communis] |
| 13 |
Hb_120117_010 |
0.1630929089 |
- |
- |
Disease resistance protein RFL1, putative [Ricinus communis] |
| 14 |
Hb_038718_010 |
0.1652336723 |
- |
- |
Disease resistance protein RFL1, putative [Ricinus communis] |
| 15 |
Hb_000692_030 |
0.1667936635 |
- |
- |
nucleoredoxin, putative [Ricinus communis] |
| 16 |
Hb_000720_060 |
0.1676469206 |
- |
- |
PREDICTED: BON1-associated protein 2-like [Jatropha curcas] |
| 17 |
Hb_106728_020 |
0.1696052936 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 18 |
Hb_000032_340 |
0.1709712954 |
- |
- |
PREDICTED: uncharacterized protein LOC105645669 [Jatropha curcas] |
| 19 |
Hb_035006_010 |
0.1710391558 |
- |
- |
hypothetical protein POPTR_0017s11350g [Populus trichocarpa] |
| 20 |
Hb_042491_010 |
0.1713142027 |
- |
- |
JHL06P13.14 [Jatropha curcas] |