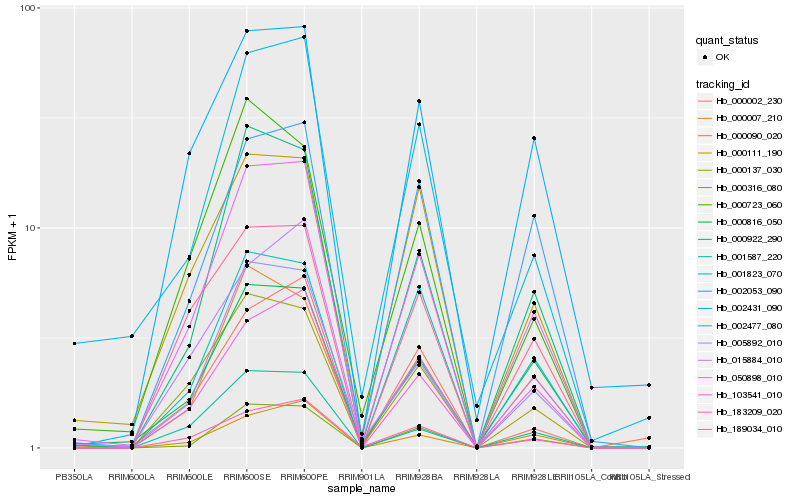
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_050898_010 |
0.0 |
- |
- |
PREDICTED: receptor-like protein kinase FERONIA [Jatropha curcas] |
| 2 |
Hb_005892_010 |
0.0763627692 |
- |
- |
PREDICTED: uncharacterized protein LOC105632168 [Jatropha curcas] |
| 3 |
Hb_000922_290 |
0.0850722593 |
- |
- |
PREDICTED: probable boron transporter 2 [Jatropha curcas] |
| 4 |
Hb_183209_020 |
0.093455872 |
- |
- |
PREDICTED: probable leucine-rich repeat receptor-like protein kinase At5g63930 isoform X2 [Jatropha curcas] |
| 5 |
Hb_000137_030 |
0.0940130774 |
- |
- |
PREDICTED: cysteine-rich repeat secretory protein 55-like [Citrus sinensis] |
| 6 |
Hb_103541_010 |
0.0942361765 |
- |
- |
PREDICTED: probable leucine-rich repeat receptor-like protein kinase At5g63930 isoform X2 [Jatropha curcas] |
| 7 |
Hb_000816_050 |
0.0994875498 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: disease resistance protein RGA2-like [Jatropha curcas] |
| 8 |
Hb_000090_020 |
0.1154362843 |
- |
- |
acyltransferase, putative [Ricinus communis] |
| 9 |
Hb_189034_010 |
0.1164318747 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 10 |
Hb_002431_090 |
0.116842047 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_001587_220 |
0.1186003209 |
- |
- |
receptor protein kinase, putative [Ricinus communis] |
| 12 |
Hb_002477_080 |
0.1197992918 |
- |
- |
hypothetical protein RCOM_0653450 [Ricinus communis] |
| 13 |
Hb_000007_210 |
0.1203613687 |
- |
- |
hypothetical protein POPTR_0010s14360g [Populus trichocarpa] |
| 14 |
Hb_001823_070 |
0.1206213522 |
- |
- |
PREDICTED: uncharacterized protein LOC105637371 [Jatropha curcas] |
| 15 |
Hb_000316_080 |
0.123359438 |
- |
- |
ENTH/VHS family protein [Theobroma cacao] |
| 16 |
Hb_002053_090 |
0.1249912539 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_000002_230 |
0.1269140796 |
- |
- |
RING/U-box superfamily protein [Theobroma cacao] |
| 18 |
Hb_015884_010 |
0.1286875203 |
- |
- |
PREDICTED: probable leucine-rich repeat receptor-like protein kinase At5g63930 isoform X1 [Jatropha curcas] |
| 19 |
Hb_000111_190 |
0.129646193 |
transcription factor |
TF Family: Trihelix |
PREDICTED: trihelix transcription factor ASIL1 [Jatropha curcas] |
| 20 |
Hb_000723_060 |
0.1318317026 |
- |
- |
PREDICTED: uncharacterized protein LOC105642243 [Jatropha curcas] |