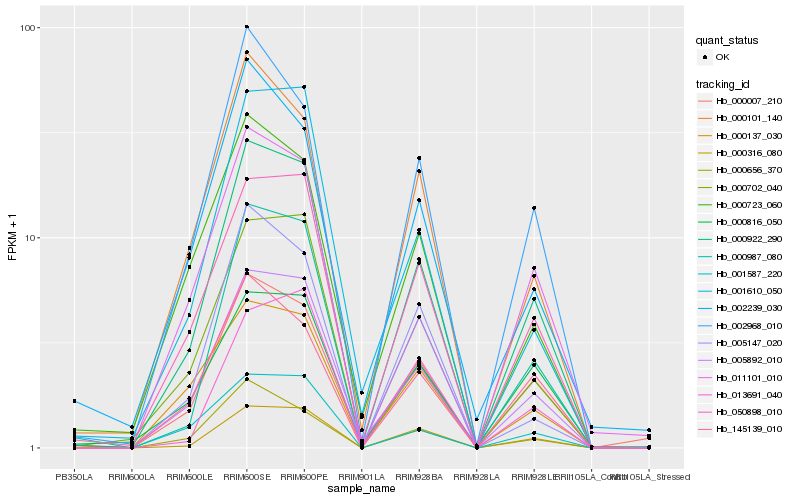
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000922_290 |
0.0 |
- |
- |
PREDICTED: probable boron transporter 2 [Jatropha curcas] |
| 2 |
Hb_005892_010 |
0.0630862305 |
- |
- |
PREDICTED: uncharacterized protein LOC105632168 [Jatropha curcas] |
| 3 |
Hb_000987_080 |
0.0808429799 |
- |
- |
light-inducible protein atls1, putative [Ricinus communis] |
| 4 |
Hb_050898_010 |
0.0850722593 |
- |
- |
PREDICTED: receptor-like protein kinase FERONIA [Jatropha curcas] |
| 5 |
Hb_000007_210 |
0.0919649684 |
- |
- |
hypothetical protein POPTR_0010s14360g [Populus trichocarpa] |
| 6 |
Hb_000656_370 |
0.1168591797 |
- |
- |
Protein kinase APK1B, chloroplast precursor, putative [Ricinus communis] |
| 7 |
Hb_145139_010 |
0.11708832 |
- |
- |
PREDICTED: sugar transporter ERD6-like 5 isoform X1 [Vitis vinifera] |
| 8 |
Hb_002968_010 |
0.118943329 |
- |
- |
PREDICTED: pectinesterase [Jatropha curcas] |
| 9 |
Hb_000316_080 |
0.120740608 |
- |
- |
ENTH/VHS family protein [Theobroma cacao] |
| 10 |
Hb_000101_140 |
0.1220017182 |
- |
- |
amino acid transporter, putative [Ricinus communis] |
| 11 |
Hb_000816_050 |
0.1263614103 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: disease resistance protein RGA2-like [Jatropha curcas] |
| 12 |
Hb_001587_220 |
0.1266381384 |
- |
- |
receptor protein kinase, putative [Ricinus communis] |
| 13 |
Hb_000137_030 |
0.1274636956 |
- |
- |
PREDICTED: cysteine-rich repeat secretory protein 55-like [Citrus sinensis] |
| 14 |
Hb_000702_040 |
0.1286344745 |
- |
- |
eukaryotic translation initiation factor 2c, putative [Ricinus communis] |
| 15 |
Hb_001610_050 |
0.1290349289 |
- |
- |
PREDICTED: subtilisin-like protease-like [Solanum tuberosum] |
| 16 |
Hb_011101_010 |
0.1300761546 |
- |
- |
hypothetical protein JCGZ_25132 [Jatropha curcas] |
| 17 |
Hb_000723_060 |
0.1310238913 |
- |
- |
PREDICTED: uncharacterized protein LOC105642243 [Jatropha curcas] |
| 18 |
Hb_005147_020 |
0.1396946175 |
- |
- |
PREDICTED: uncharacterized protein LOC105629671 [Jatropha curcas] |
| 19 |
Hb_002239_030 |
0.1420131719 |
transcription factor |
TF Family: ARF |
PREDICTED: auxin response factor 4 [Jatropha curcas] |
| 20 |
Hb_013691_040 |
0.1449382237 |
- |
- |
hypothetical protein B456_001G045900 [Gossypium raimondii] |