| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
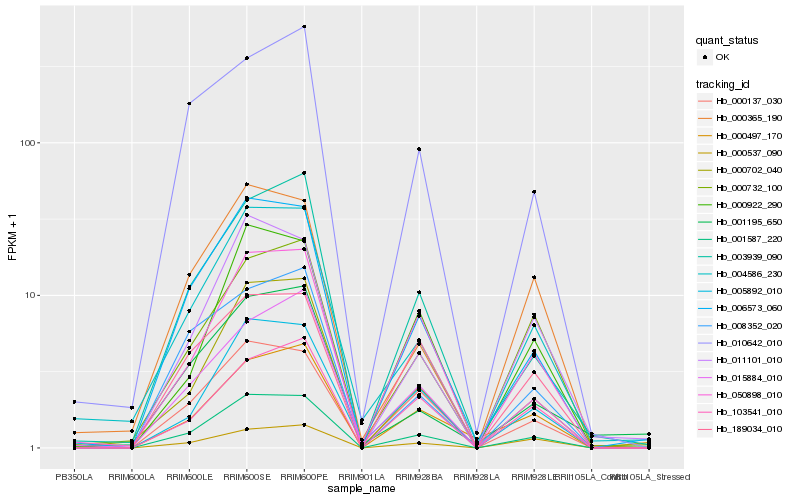
Hb_001587_220 |
0.0 |
- |
- |
receptor protein kinase, putative [Ricinus communis] |
| 2 |
Hb_006573_060 |
0.0706388606 |
- |
- |
hypothetical protein [Bruguiera gymnorhiza] |
| 3 |
Hb_000702_040 |
0.1029159402 |
- |
- |
eukaryotic translation initiation factor 2c, putative [Ricinus communis] |
| 4 |
Hb_005892_010 |
0.1078096569 |
- |
- |
PREDICTED: uncharacterized protein LOC105632168 [Jatropha curcas] |
| 5 |
Hb_011101_010 |
0.108394239 |
- |
- |
hypothetical protein JCGZ_25132 [Jatropha curcas] |
| 6 |
Hb_004586_230 |
0.1089858113 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_000137_030 |
0.1138735529 |
- |
- |
PREDICTED: cysteine-rich repeat secretory protein 55-like [Citrus sinensis] |
| 8 |
Hb_050898_010 |
0.1186003209 |
- |
- |
PREDICTED: receptor-like protein kinase FERONIA [Jatropha curcas] |
| 9 |
Hb_003939_090 |
0.1256601118 |
transcription factor |
TF Family: NAC |
transcription factor, putative [Ricinus communis] |
| 10 |
Hb_000922_290 |
0.1266381384 |
- |
- |
PREDICTED: probable boron transporter 2 [Jatropha curcas] |
| 11 |
Hb_015884_010 |
0.1291321945 |
- |
- |
PREDICTED: probable leucine-rich repeat receptor-like protein kinase At5g63930 isoform X1 [Jatropha curcas] |
| 12 |
Hb_000365_190 |
0.1308441816 |
- |
- |
PREDICTED: protein LURP-one-related 8 [Jatropha curcas] |
| 13 |
Hb_000497_170 |
0.131110671 |
- |
- |
PREDICTED: uncharacterized protein LOC105647275 [Jatropha curcas] |
| 14 |
Hb_000732_100 |
0.1322219934 |
- |
- |
PREDICTED: uncharacterized protein LOC105649440 [Jatropha curcas] |
| 15 |
Hb_001195_650 |
0.1366303284 |
- |
- |
PREDICTED: calcium uniporter protein 4, mitochondrial [Jatropha curcas] |
| 16 |
Hb_008352_020 |
0.136763926 |
- |
- |
hypothetical protein [Cecembia lonarensis] |
| 17 |
Hb_189034_010 |
0.1380110957 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 18 |
Hb_000537_090 |
0.1386954074 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_103541_010 |
0.1409491578 |
- |
- |
PREDICTED: probable leucine-rich repeat receptor-like protein kinase At5g63930 isoform X2 [Jatropha curcas] |
| 20 |
Hb_010642_010 |
0.1413618652 |
- |
- |
PREDICTED: aquaporin TIP2-1 [Jatropha curcas] |