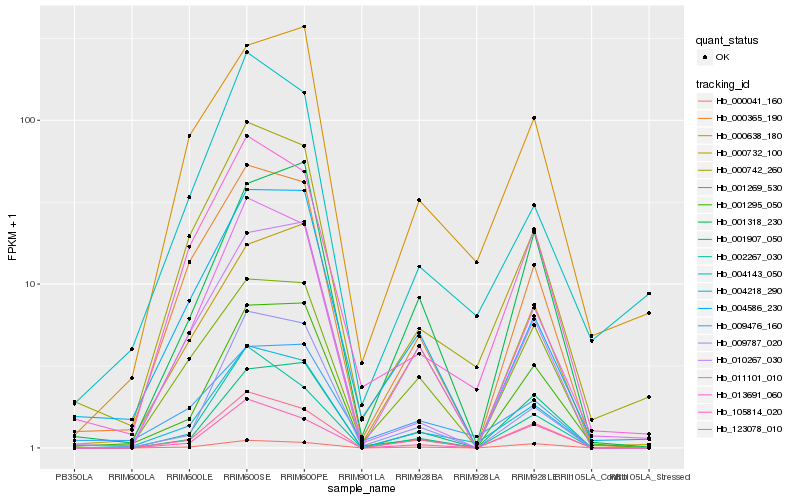
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004218_290 |
0.0 |
transcription factor |
TF Family: SRS |
PREDICTED: protein SHI RELATED SEQUENCE 6 isoform X2 [Jatropha curcas] |
| 2 |
Hb_011101_010 |
0.1081580217 |
- |
- |
hypothetical protein JCGZ_25132 [Jatropha curcas] |
| 3 |
Hb_123078_010 |
0.1096614193 |
- |
- |
PREDICTED: LRR receptor-like serine/threonine-protein kinase EFR [Jatropha curcas] |
| 4 |
Hb_000742_260 |
0.1120206989 |
- |
- |
hypothetical protein RCOM_1336320 [Ricinus communis] |
| 5 |
Hb_000365_190 |
0.1193717658 |
- |
- |
PREDICTED: protein LURP-one-related 8 [Jatropha curcas] |
| 6 |
Hb_013691_060 |
0.1277769443 |
- |
- |
PREDICTED: uncharacterized protein LOC105119066 [Populus euphratica] |
| 7 |
Hb_002267_030 |
0.1384165448 |
- |
- |
hypothetical protein JCGZ_24883 [Jatropha curcas] |
| 8 |
Hb_001269_530 |
0.1448544615 |
- |
- |
ATP-binding cassette transporter, putative [Ricinus communis] |
| 9 |
Hb_105814_020 |
0.1452518587 |
- |
- |
berberine bridge enzyme [Hevea brasiliensis] |
| 10 |
Hb_000041_160 |
0.1464968715 |
- |
- |
PREDICTED: uncharacterized protein LOC104907627 [Beta vulgaris subsp. vulgaris] |
| 11 |
Hb_009787_020 |
0.146570982 |
- |
- |
PREDICTED: cytochrome P450 705A5-like [Jatropha curcas] |
| 12 |
Hb_001295_050 |
0.1465796559 |
- |
- |
PREDICTED: uncharacterized protein LOC101503108 isoform X1 [Cicer arietinum] |
| 13 |
Hb_000638_180 |
0.1473961988 |
- |
- |
PREDICTED: uncharacterized protein LOC105124685 [Populus euphratica] |
| 14 |
Hb_010267_030 |
0.1525312289 |
- |
- |
DUF26 domain-containing protein 1 precursor, putative [Ricinus communis] |
| 15 |
Hb_000732_100 |
0.1535265547 |
- |
- |
PREDICTED: uncharacterized protein LOC105649440 [Jatropha curcas] |
| 16 |
Hb_009476_160 |
0.15421913 |
- |
- |
RING-H2 finger protein ATL2N, putative [Ricinus communis] |
| 17 |
Hb_001318_230 |
0.1545186468 |
- |
- |
Boron transporter, putative [Ricinus communis] |
| 18 |
Hb_004586_230 |
0.1565608856 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_004143_050 |
0.156779861 |
- |
- |
PREDICTED: uncharacterized protein LOC105630712 [Jatropha curcas] |
| 20 |
Hb_001907_050 |
0.1570751213 |
- |
- |
Receptor like protein 6, putative [Theobroma cacao] |