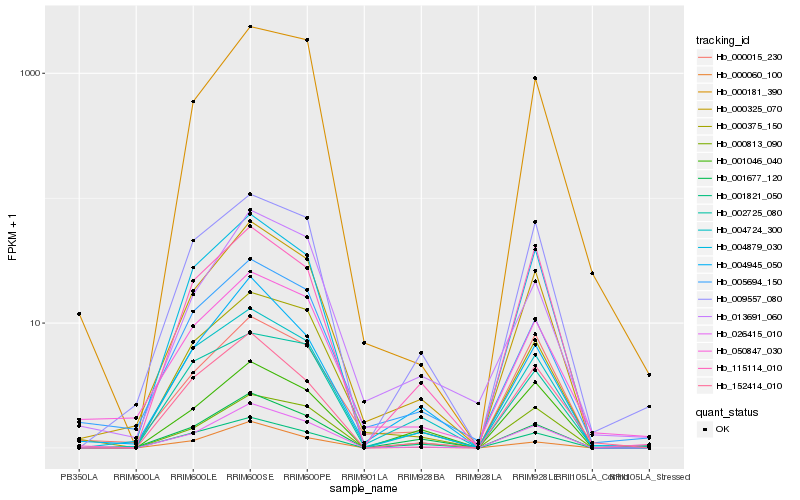
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000325_070 |
0.0 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |
| 2 |
Hb_026415_010 |
0.0730041986 |
- |
- |
PREDICTED: probable receptor-like protein kinase At1g67000 isoform X1 [Jatropha curcas] |
| 3 |
Hb_005694_150 |
0.0959544328 |
- |
- |
PREDICTED: probable mitochondrial chaperone BCS1-A [Jatropha curcas] |
| 4 |
Hb_000375_150 |
0.0960797547 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_004879_030 |
0.0999015979 |
- |
- |
PREDICTED: uncharacterized protein LOC105124685 [Populus euphratica] |
| 6 |
Hb_004724_300 |
0.1005641202 |
transcription factor |
TF Family: NAC |
PREDICTED: NAC domain-containing protein 90-like [Jatropha curcas] |
| 7 |
Hb_000015_230 |
0.1048461211 |
transcription factor |
TF Family: C2C2-Dof |
PREDICTED: dof zinc finger protein DOF3.6-like isoform X2 [Populus euphratica] |
| 8 |
Hb_004945_050 |
0.1073296233 |
transcription factor |
TF Family: C2C2-Dof |
hypothetical protein POPTR_0008s08740g [Populus trichocarpa] |
| 9 |
Hb_009557_080 |
0.1101206134 |
- |
- |
PREDICTED: glutaredoxin-C9-like [Populus euphratica] |
| 10 |
Hb_050847_030 |
0.1123768044 |
- |
- |
PREDICTED: cytochrome P450 714A1-like [Populus euphratica] |
| 11 |
Hb_001677_120 |
0.1157316125 |
- |
- |
PREDICTED: receptor-like protein 12 [Jatropha curcas] |
| 12 |
Hb_013691_060 |
0.1175946174 |
- |
- |
PREDICTED: uncharacterized protein LOC105119066 [Populus euphratica] |
| 13 |
Hb_000813_090 |
0.1209736507 |
- |
- |
hypothetical protein CARUB_v10019318mg [Capsella rubella] |
| 14 |
Hb_115114_010 |
0.1232036083 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_002725_080 |
0.126396674 |
- |
- |
protein phosphatase 2c, putative [Ricinus communis] |
| 16 |
Hb_001821_050 |
0.1280120681 |
- |
- |
hypothetical protein CICLE_v10032084mg [Citrus clementina] |
| 17 |
Hb_000181_390 |
0.1296977694 |
- |
- |
unknown [Lotus japonicus] |
| 18 |
Hb_000060_100 |
0.1311907026 |
transcription factor |
TF Family: OFP |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_152414_010 |
0.1316333108 |
desease resistance |
Gene Name: NB-ARC |
hypothetical protein PRUPE_ppa024232mg [Prunus persica] |
| 20 |
Hb_001046_040 |
0.1358894 |
- |
- |
UDP-glucosyltransferase, putative [Ricinus communis] |