| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
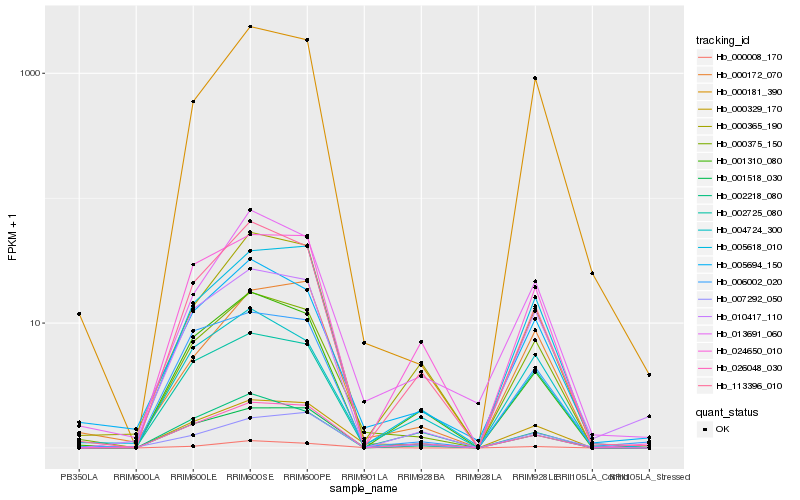
Hb_000329_170 |
0.0 |
- |
- |
PREDICTED: cysteine-rich repeat secretory protein 15 [Jatropha curcas] |
| 2 |
Hb_000375_150 |
0.0921230112 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_001518_030 |
0.1044577443 |
- |
- |
Receptor protein kinase CLAVATA1 precursor, putative [Ricinus communis] |
| 4 |
Hb_002725_080 |
0.1123476559 |
- |
- |
protein phosphatase 2c, putative [Ricinus communis] |
| 5 |
Hb_005618_010 |
0.1162270302 |
transcription factor |
TF Family: MYB |
hypothetical protein POPTR_0014s10680g [Populus trichocarpa] |
| 6 |
Hb_026048_030 |
0.1288067605 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_024650_010 |
0.1291980089 |
- |
- |
PREDICTED: BAHD acyltransferase DCR [Jatropha curcas] |
| 8 |
Hb_113396_010 |
0.1393317283 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 9 |
Hb_000008_170 |
0.1421758168 |
- |
- |
PREDICTED: uncharacterized protein LOC105628076 [Jatropha curcas] |
| 10 |
Hb_005694_150 |
0.1422990767 |
- |
- |
PREDICTED: probable mitochondrial chaperone BCS1-A [Jatropha curcas] |
| 11 |
Hb_006002_020 |
0.1427519678 |
- |
- |
PREDICTED: cytokinin dehydrogenase 7 [Jatropha curcas] |
| 12 |
Hb_004724_300 |
0.1432707151 |
transcription factor |
TF Family: NAC |
PREDICTED: NAC domain-containing protein 90-like [Jatropha curcas] |
| 13 |
Hb_000365_190 |
0.1433337625 |
- |
- |
PREDICTED: protein LURP-one-related 8 [Jatropha curcas] |
| 14 |
Hb_007292_050 |
0.1444724866 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: disease resistance RPP13-like protein 4 [Jatropha curcas] |
| 15 |
Hb_001310_080 |
0.1445212602 |
transcription factor |
TF Family: C2H2 |
PREDICTED: zinc finger protein AZF2-like [Jatropha curcas] |
| 16 |
Hb_000172_070 |
0.1445812888 |
- |
- |
trehalose-6-phosphate synthase, putative [Ricinus communis] |
| 17 |
Hb_010417_110 |
0.1453738433 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_013691_060 |
0.1470907225 |
- |
- |
PREDICTED: uncharacterized protein LOC105119066 [Populus euphratica] |
| 19 |
Hb_002218_080 |
0.147879486 |
- |
- |
Ankyrin repeat family protein, putative [Theobroma cacao] |
| 20 |
Hb_000181_390 |
0.1491461199 |
- |
- |
unknown [Lotus japonicus] |