| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
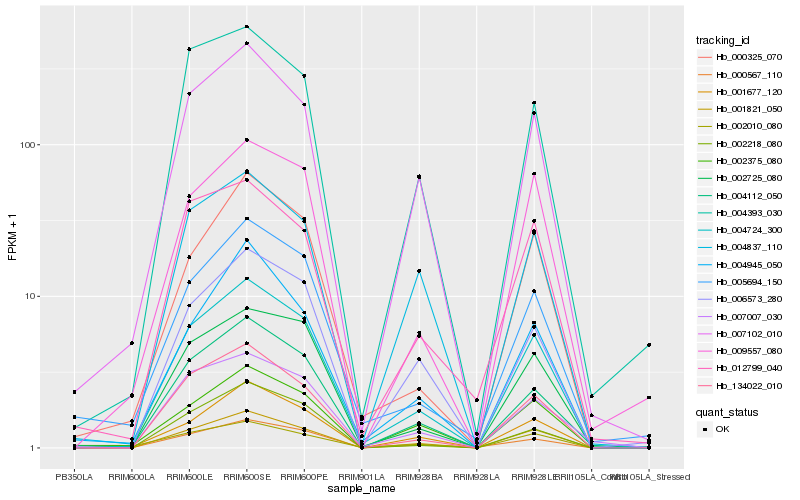
Hb_004724_300 |
0.0 |
transcription factor |
TF Family: NAC |
PREDICTED: NAC domain-containing protein 90-like [Jatropha curcas] |
| 2 |
Hb_001821_050 |
0.0581409185 |
- |
- |
hypothetical protein CICLE_v10032084mg [Citrus clementina] |
| 3 |
Hb_134022_010 |
0.0802940157 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: disease resistance protein RPM1 [Jatropha curcas] |
| 4 |
Hb_002725_080 |
0.0842423984 |
- |
- |
protein phosphatase 2c, putative [Ricinus communis] |
| 5 |
Hb_007102_010 |
0.0907215317 |
- |
- |
Granule-bound starch synthase 1, chloroplastic/amyloplastic [Gossypium arboreum] |
| 6 |
Hb_004112_050 |
0.093440604 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance protein RGA3 [Jatropha curcas] |
| 7 |
Hb_001677_120 |
0.0958846616 |
- |
- |
PREDICTED: receptor-like protein 12 [Jatropha curcas] |
| 8 |
Hb_000325_070 |
0.1005641202 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |
| 9 |
Hb_009557_080 |
0.106736428 |
- |
- |
PREDICTED: glutaredoxin-C9-like [Populus euphratica] |
| 10 |
Hb_006573_280 |
0.1070271578 |
- |
- |
PREDICTED: probable membrane-associated kinase regulator 5 [Jatropha curcas] |
| 11 |
Hb_000567_110 |
0.1083430297 |
- |
- |
hypothetical protein JCGZ_21556 [Jatropha curcas] |
| 12 |
Hb_002010_080 |
0.1117281068 |
- |
- |
PREDICTED: probable receptor-like protein kinase At1g11050 [Jatropha curcas] |
| 13 |
Hb_004393_030 |
0.111811129 |
- |
- |
CBL-interacting protein kinase 6 [Populus trichocarpa] |
| 14 |
Hb_002218_080 |
0.1138438631 |
- |
- |
Ankyrin repeat family protein, putative [Theobroma cacao] |
| 15 |
Hb_005694_150 |
0.113918471 |
- |
- |
PREDICTED: probable mitochondrial chaperone BCS1-A [Jatropha curcas] |
| 16 |
Hb_007007_030 |
0.1140358198 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_004837_110 |
0.1183685725 |
- |
- |
PREDICTED: MATE efflux family protein 1 isoform X1 [Jatropha curcas] |
| 18 |
Hb_004945_050 |
0.1184047461 |
transcription factor |
TF Family: C2C2-Dof |
hypothetical protein POPTR_0008s08740g [Populus trichocarpa] |
| 19 |
Hb_002375_080 |
0.1202143005 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_012799_040 |
0.1206471061 |
- |
- |
PREDICTED: transmembrane ascorbate ferrireductase 1 [Jatropha curcas] |