| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
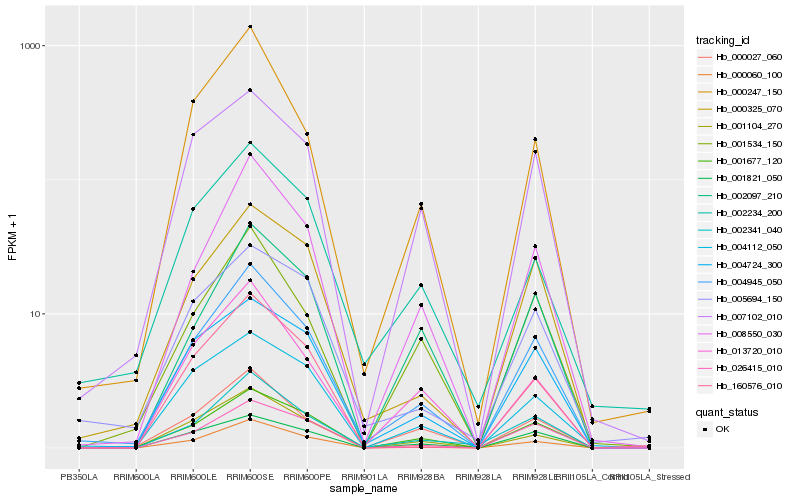
Hb_004945_050 |
0.0 |
transcription factor |
TF Family: C2C2-Dof |
hypothetical protein POPTR_0008s08740g [Populus trichocarpa] |
| 2 |
Hb_000060_100 |
0.0937226919 |
transcription factor |
TF Family: OFP |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_001677_120 |
0.0968324163 |
- |
- |
PREDICTED: receptor-like protein 12 [Jatropha curcas] |
| 4 |
Hb_008550_030 |
0.0978083171 |
- |
- |
PREDICTED: tonoplast dicarboxylate transporter [Jatropha curcas] |
| 5 |
Hb_002341_040 |
0.1007208716 |
- |
- |
serine/threonine-protein kinase bri1, putative [Ricinus communis] |
| 6 |
Hb_000325_070 |
0.1073296233 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |
| 7 |
Hb_000027_060 |
0.117629433 |
- |
- |
hypothetical protein POPTR_0004s07470g [Populus trichocarpa] |
| 8 |
Hb_004724_300 |
0.1184047461 |
transcription factor |
TF Family: NAC |
PREDICTED: NAC domain-containing protein 90-like [Jatropha curcas] |
| 9 |
Hb_001104_270 |
0.1203078621 |
- |
- |
hypothetical protein POPTR_0007s13980g [Populus trichocarpa] |
| 10 |
Hb_000247_150 |
0.1224666007 |
- |
- |
PREDICTED: haloacid dehalogenase-like hydrolase domain-containing protein 3 [Jatropha curcas] |
| 11 |
Hb_001821_050 |
0.1244970927 |
- |
- |
hypothetical protein CICLE_v10032084mg [Citrus clementina] |
| 12 |
Hb_001534_150 |
0.1257808235 |
- |
- |
PREDICTED: uncharacterized permease C29B12.14c [Jatropha curcas] |
| 13 |
Hb_007102_010 |
0.1259072156 |
- |
- |
Granule-bound starch synthase 1, chloroplastic/amyloplastic [Gossypium arboreum] |
| 14 |
Hb_004112_050 |
0.1271893086 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance protein RGA3 [Jatropha curcas] |
| 15 |
Hb_013720_010 |
0.129206043 |
- |
- |
Putative DNA repair RAD23-3 -like protein [Gossypium arboreum] |
| 16 |
Hb_002097_210 |
0.1319880523 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_005694_150 |
0.1321559605 |
- |
- |
PREDICTED: probable mitochondrial chaperone BCS1-A [Jatropha curcas] |
| 18 |
Hb_026415_010 |
0.1329245017 |
- |
- |
PREDICTED: probable receptor-like protein kinase At1g67000 isoform X1 [Jatropha curcas] |
| 19 |
Hb_160576_010 |
0.1366554111 |
- |
- |
PREDICTED: probable receptor-like protein kinase At1g67000 isoform X1 [Jatropha curcas] |
| 20 |
Hb_002234_200 |
0.1387653352 |
- |
- |
PREDICTED: inactive rhomboid protein 1 [Jatropha curcas] |