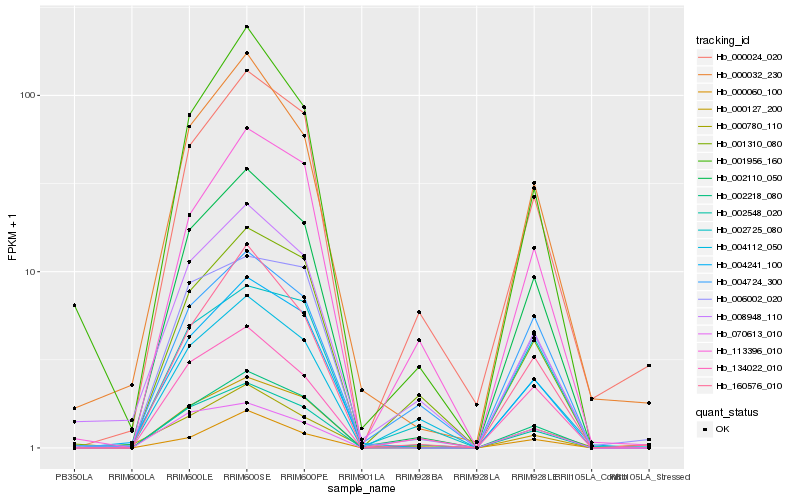
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002110_050 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_002548_020 |
0.0550068091 |
- |
- |
PREDICTED: probable receptor-like protein kinase At1g67000 isoform X1 [Jatropha curcas] |
| 3 |
Hb_002218_080 |
0.0632328985 |
- |
- |
Ankyrin repeat family protein, putative [Theobroma cacao] |
| 4 |
Hb_004241_100 |
0.0777630982 |
- |
- |
PREDICTED: transcription factor bHLH123-like isoform X1 [Populus euphratica] |
| 5 |
Hb_160576_010 |
0.0820732613 |
- |
- |
PREDICTED: probable receptor-like protein kinase At1g67000 isoform X1 [Jatropha curcas] |
| 6 |
Hb_000032_230 |
0.0970127061 |
transcription factor |
TF Family: ERF |
hypothetical protein JCGZ_20463 [Jatropha curcas] |
| 7 |
Hb_134022_010 |
0.097442291 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: disease resistance protein RPM1 [Jatropha curcas] |
| 8 |
Hb_070613_010 |
0.1094523001 |
- |
- |
PREDICTED: probable receptor-like protein kinase At1g67000 [Jatropha curcas] |
| 9 |
Hb_000060_100 |
0.1162130393 |
transcription factor |
TF Family: OFP |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_000024_020 |
0.1171116578 |
- |
- |
PREDICTED: uncharacterized protein LOC105636896 [Jatropha curcas] |
| 11 |
Hb_006002_020 |
0.1171793362 |
- |
- |
PREDICTED: cytokinin dehydrogenase 7 [Jatropha curcas] |
| 12 |
Hb_001956_160 |
0.1194401115 |
- |
- |
hypothetical protein L484_009438 [Morus notabilis] |
| 13 |
Hb_001310_080 |
0.1214002385 |
transcription factor |
TF Family: C2H2 |
PREDICTED: zinc finger protein AZF2-like [Jatropha curcas] |
| 14 |
Hb_004112_050 |
0.122740012 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance protein RGA3 [Jatropha curcas] |
| 15 |
Hb_004724_300 |
0.123620411 |
transcription factor |
TF Family: NAC |
PREDICTED: NAC domain-containing protein 90-like [Jatropha curcas] |
| 16 |
Hb_113396_010 |
0.1239849454 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 17 |
Hb_008948_110 |
0.1240449506 |
- |
- |
PREDICTED: peroxidase 25 [Jatropha curcas] |
| 18 |
Hb_000780_110 |
0.1253753563 |
- |
- |
cation-transporting atpase plant, putative [Ricinus communis] |
| 19 |
Hb_000127_200 |
0.1262134762 |
- |
- |
PREDICTED: uncharacterized protein LOC100255537 [Vitis vinifera] |
| 20 |
Hb_002725_080 |
0.132128721 |
- |
- |
protein phosphatase 2c, putative [Ricinus communis] |