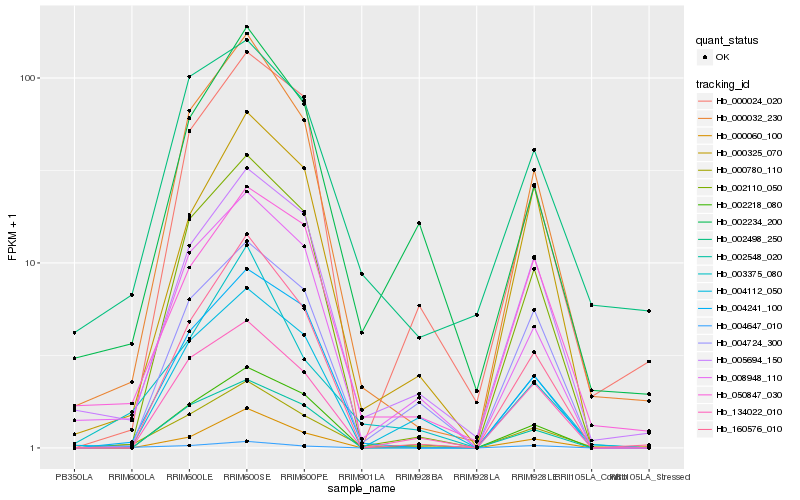
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000780_110 |
0.0 |
- |
- |
cation-transporting atpase plant, putative [Ricinus communis] |
| 2 |
Hb_000032_230 |
0.0945694848 |
transcription factor |
TF Family: ERF |
hypothetical protein JCGZ_20463 [Jatropha curcas] |
| 3 |
Hb_002110_050 |
0.1253753563 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_004241_100 |
0.1278924482 |
- |
- |
PREDICTED: transcription factor bHLH123-like isoform X1 [Populus euphratica] |
| 5 |
Hb_008948_110 |
0.1284062054 |
- |
- |
PREDICTED: peroxidase 25 [Jatropha curcas] |
| 6 |
Hb_005694_150 |
0.1311070074 |
- |
- |
PREDICTED: probable mitochondrial chaperone BCS1-A [Jatropha curcas] |
| 7 |
Hb_160576_010 |
0.1421744976 |
- |
- |
PREDICTED: probable receptor-like protein kinase At1g67000 isoform X1 [Jatropha curcas] |
| 8 |
Hb_002218_080 |
0.1452602236 |
- |
- |
Ankyrin repeat family protein, putative [Theobroma cacao] |
| 9 |
Hb_050847_030 |
0.1458421761 |
- |
- |
PREDICTED: cytochrome P450 714A1-like [Populus euphratica] |
| 10 |
Hb_134022_010 |
0.1465059634 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: disease resistance protein RPM1 [Jatropha curcas] |
| 11 |
Hb_004112_050 |
0.1474171952 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance protein RGA3 [Jatropha curcas] |
| 12 |
Hb_000325_070 |
0.1493097037 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |
| 13 |
Hb_004724_300 |
0.1502191722 |
transcription factor |
TF Family: NAC |
PREDICTED: NAC domain-containing protein 90-like [Jatropha curcas] |
| 14 |
Hb_002498_250 |
0.1510141234 |
- |
- |
5'-adenylylsulfate reductase 1, chloroplast precursor, putative [Ricinus communis] |
| 15 |
Hb_003375_080 |
0.1517817281 |
- |
- |
PREDICTED: uncharacterized protein LOC105643382 isoform X1 [Jatropha curcas] |
| 16 |
Hb_002548_020 |
0.1526050259 |
- |
- |
PREDICTED: probable receptor-like protein kinase At1g67000 isoform X1 [Jatropha curcas] |
| 17 |
Hb_002234_200 |
0.1528154016 |
- |
- |
PREDICTED: inactive rhomboid protein 1 [Jatropha curcas] |
| 18 |
Hb_000060_100 |
0.1529955989 |
transcription factor |
TF Family: OFP |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_004647_010 |
0.1556913463 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 20 |
Hb_000024_020 |
0.1566791898 |
- |
- |
PREDICTED: uncharacterized protein LOC105636896 [Jatropha curcas] |