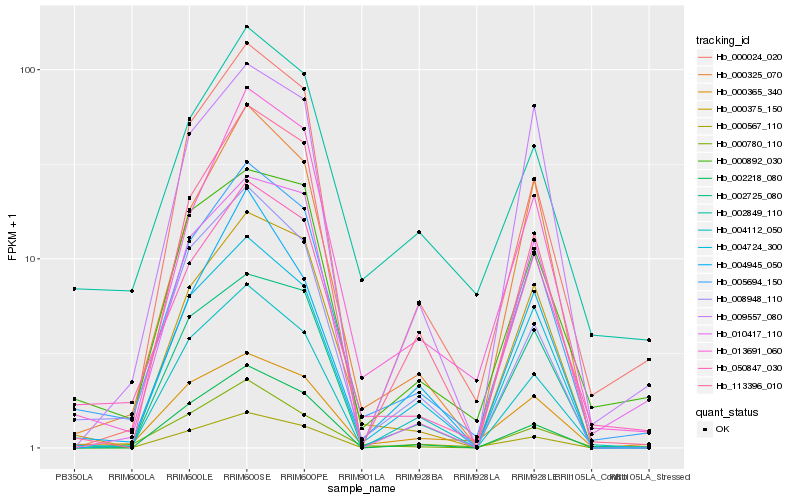
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005694_150 |
0.0 |
- |
- |
PREDICTED: probable mitochondrial chaperone BCS1-A [Jatropha curcas] |
| 2 |
Hb_050847_030 |
0.0633681997 |
- |
- |
PREDICTED: cytochrome P450 714A1-like [Populus euphratica] |
| 3 |
Hb_000375_150 |
0.0941712123 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_000325_070 |
0.0959544328 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |
| 5 |
Hb_000365_340 |
0.1050831492 |
- |
- |
PREDICTED: uncharacterized protein LOC105649067 [Jatropha curcas] |
| 6 |
Hb_013691_060 |
0.1063437431 |
- |
- |
PREDICTED: uncharacterized protein LOC105119066 [Populus euphratica] |
| 7 |
Hb_000892_030 |
0.107029244 |
- |
- |
PREDICTED: uncharacterized protein At1g04910 [Jatropha curcas] |
| 8 |
Hb_004724_300 |
0.113918471 |
transcription factor |
TF Family: NAC |
PREDICTED: NAC domain-containing protein 90-like [Jatropha curcas] |
| 9 |
Hb_000567_110 |
0.1154841673 |
- |
- |
hypothetical protein JCGZ_21556 [Jatropha curcas] |
| 10 |
Hb_002725_080 |
0.1169490681 |
- |
- |
protein phosphatase 2c, putative [Ricinus communis] |
| 11 |
Hb_000024_020 |
0.1197040887 |
- |
- |
PREDICTED: uncharacterized protein LOC105636896 [Jatropha curcas] |
| 12 |
Hb_008948_110 |
0.1205178687 |
- |
- |
PREDICTED: peroxidase 25 [Jatropha curcas] |
| 13 |
Hb_004112_050 |
0.1211486964 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance protein RGA3 [Jatropha curcas] |
| 14 |
Hb_002849_110 |
0.1254165547 |
- |
- |
hypothetical protein JCGZ_14362 [Jatropha curcas] |
| 15 |
Hb_113396_010 |
0.1257838176 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 16 |
Hb_000780_110 |
0.1311070074 |
- |
- |
cation-transporting atpase plant, putative [Ricinus communis] |
| 17 |
Hb_010417_110 |
0.1317180178 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_004945_050 |
0.1321559605 |
transcription factor |
TF Family: C2C2-Dof |
hypothetical protein POPTR_0008s08740g [Populus trichocarpa] |
| 19 |
Hb_009557_080 |
0.1331030779 |
- |
- |
PREDICTED: glutaredoxin-C9-like [Populus euphratica] |
| 20 |
Hb_002218_080 |
0.1345722465 |
- |
- |
Ankyrin repeat family protein, putative [Theobroma cacao] |