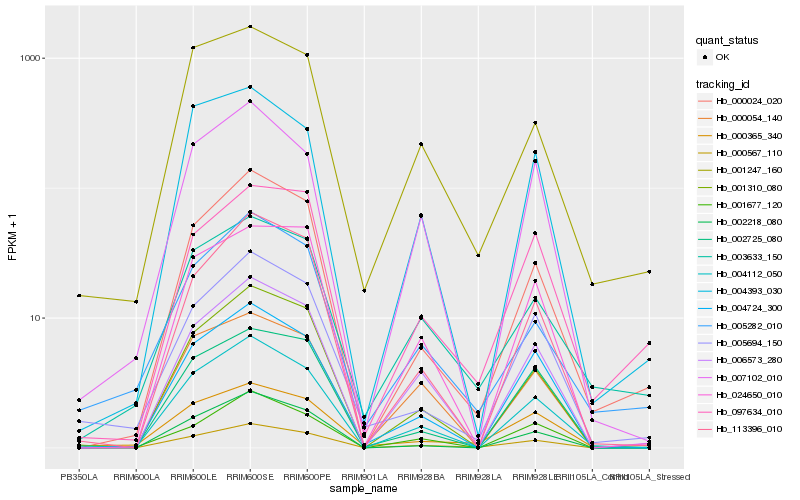
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000567_110 |
0.0 |
- |
- |
hypothetical protein JCGZ_21556 [Jatropha curcas] |
| 2 |
Hb_000024_020 |
0.1014994519 |
- |
- |
PREDICTED: uncharacterized protein LOC105636896 [Jatropha curcas] |
| 3 |
Hb_006573_280 |
0.1061269265 |
- |
- |
PREDICTED: probable membrane-associated kinase regulator 5 [Jatropha curcas] |
| 4 |
Hb_004724_300 |
0.1083430297 |
transcription factor |
TF Family: NAC |
PREDICTED: NAC domain-containing protein 90-like [Jatropha curcas] |
| 5 |
Hb_003633_150 |
0.1112691774 |
- |
- |
hypothetical protein JCGZ_23296 [Jatropha curcas] |
| 6 |
Hb_004112_050 |
0.1131998941 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance protein RGA3 [Jatropha curcas] |
| 7 |
Hb_005694_150 |
0.1154841673 |
- |
- |
PREDICTED: probable mitochondrial chaperone BCS1-A [Jatropha curcas] |
| 8 |
Hb_001247_160 |
0.117250724 |
- |
- |
s-adenosylmethionine synthetase, putative [Ricinus communis] |
| 9 |
Hb_000365_340 |
0.1199615997 |
- |
- |
PREDICTED: uncharacterized protein LOC105649067 [Jatropha curcas] |
| 10 |
Hb_001310_080 |
0.124070465 |
transcription factor |
TF Family: C2H2 |
PREDICTED: zinc finger protein AZF2-like [Jatropha curcas] |
| 11 |
Hb_097634_010 |
0.1251489273 |
- |
- |
PREDICTED: UDP-glucuronate 4-epimerase 1 [Jatropha curcas] |
| 12 |
Hb_002725_080 |
0.1267420883 |
- |
- |
protein phosphatase 2c, putative [Ricinus communis] |
| 13 |
Hb_004393_030 |
0.1271608824 |
- |
- |
CBL-interacting protein kinase 6 [Populus trichocarpa] |
| 14 |
Hb_005282_010 |
0.127349755 |
- |
- |
PREDICTED: probable xyloglucan glycosyltransferase 12 [Populus euphratica] |
| 15 |
Hb_000054_140 |
0.1279458153 |
- |
- |
PREDICTED: MACPF domain-containing protein CAD1 [Jatropha curcas] |
| 16 |
Hb_001677_120 |
0.1280662535 |
- |
- |
PREDICTED: receptor-like protein 12 [Jatropha curcas] |
| 17 |
Hb_113396_010 |
0.1281259451 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 18 |
Hb_007102_010 |
0.1282364169 |
- |
- |
Granule-bound starch synthase 1, chloroplastic/amyloplastic [Gossypium arboreum] |
| 19 |
Hb_002218_080 |
0.1320665049 |
- |
- |
Ankyrin repeat family protein, putative [Theobroma cacao] |
| 20 |
Hb_024650_010 |
0.1324327085 |
- |
- |
PREDICTED: BAHD acyltransferase DCR [Jatropha curcas] |