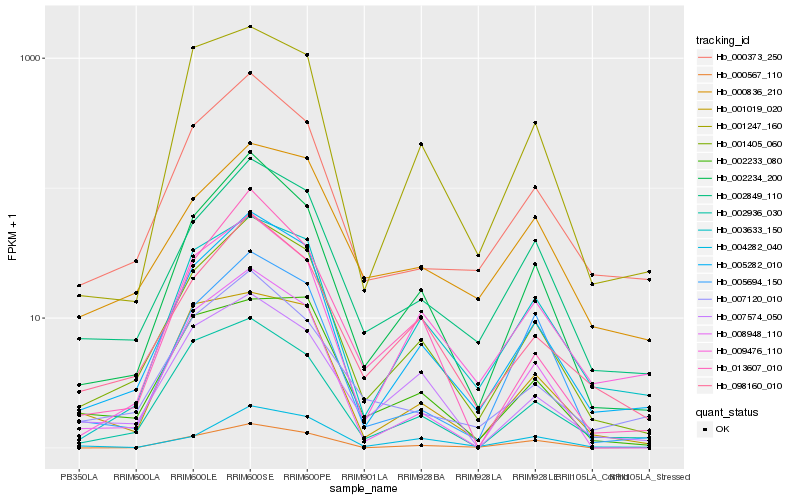
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001405_060 |
0.0 |
- |
- |
PREDICTED: CBL-interacting serine/threonine-protein kinase 4-like [Jatropha curcas] |
| 2 |
Hb_005282_010 |
0.0553094222 |
- |
- |
PREDICTED: probable xyloglucan glycosyltransferase 12 [Populus euphratica] |
| 3 |
Hb_002234_200 |
0.0739102758 |
- |
- |
PREDICTED: inactive rhomboid protein 1 [Jatropha curcas] |
| 4 |
Hb_098160_010 |
0.0936016993 |
desease resistance |
Gene Name: TIR |
PREDICTED: TMV resistance protein N-like isoform X2 [Jatropha curcas] |
| 5 |
Hb_002849_110 |
0.0948296412 |
- |
- |
hypothetical protein JCGZ_14362 [Jatropha curcas] |
| 6 |
Hb_000373_250 |
0.1158117017 |
- |
- |
PREDICTED: uncharacterized protein LOC105640939 [Jatropha curcas] |
| 7 |
Hb_003633_150 |
0.1191448384 |
- |
- |
hypothetical protein JCGZ_23296 [Jatropha curcas] |
| 8 |
Hb_007574_050 |
0.1212472537 |
- |
- |
PREDICTED: probable leucine-rich repeat receptor-like protein kinase At1g68400 [Jatropha curcas] |
| 9 |
Hb_008948_110 |
0.1221731972 |
- |
- |
PREDICTED: peroxidase 25 [Jatropha curcas] |
| 10 |
Hb_002936_030 |
0.1259648344 |
- |
- |
PREDICTED: uncharacterized protein LOC105633479 isoform X1 [Jatropha curcas] |
| 11 |
Hb_001247_160 |
0.1315628977 |
- |
- |
s-adenosylmethionine synthetase, putative [Ricinus communis] |
| 12 |
Hb_013607_010 |
0.1358126224 |
transcription factor |
TF Family: bZIP |
PREDICTED: transcription factor TGA3 [Jatropha curcas] |
| 13 |
Hb_007120_010 |
0.1360193396 |
- |
- |
PREDICTED: uncharacterized protein LOC105634737 [Jatropha curcas] |
| 14 |
Hb_000567_110 |
0.1400081836 |
- |
- |
hypothetical protein JCGZ_21556 [Jatropha curcas] |
| 15 |
Hb_004282_040 |
0.1403152999 |
desease resistance |
Gene Name: TIR |
PREDICTED: TMV resistance protein N-like isoform X2 [Jatropha curcas] |
| 16 |
Hb_005694_150 |
0.1409554563 |
- |
- |
PREDICTED: probable mitochondrial chaperone BCS1-A [Jatropha curcas] |
| 17 |
Hb_002233_080 |
0.1421800649 |
- |
- |
hypothetical protein JCGZ_00222 [Jatropha curcas] |
| 18 |
Hb_009476_110 |
0.1424134806 |
- |
- |
PREDICTED: probable protein phosphatase 2C 25 [Jatropha curcas] |
| 19 |
Hb_000836_210 |
0.1440354015 |
- |
- |
hypothetical protein JCGZ_14362 [Jatropha curcas] |
| 20 |
Hb_001019_020 |
0.1446422203 |
- |
- |
conserved hypothetical protein [Ricinus communis] |