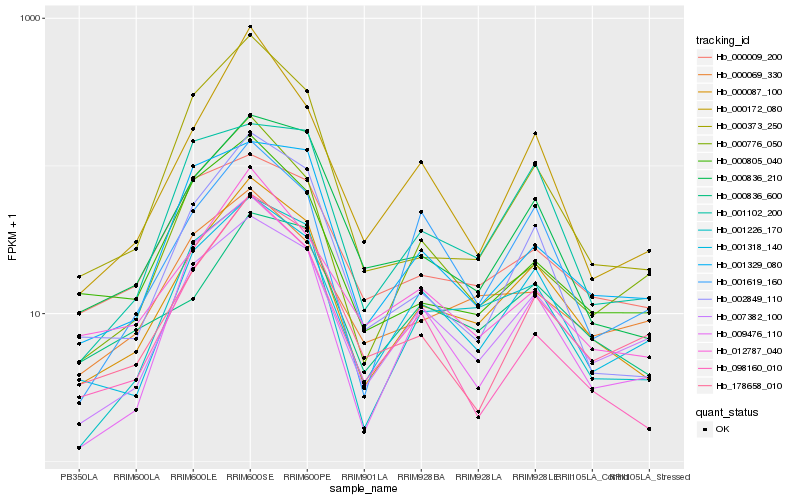
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000087_100 |
0.0 |
- |
- |
PREDICTED: cellulose synthase-like protein D3 [Jatropha curcas] |
| 2 |
Hb_001318_140 |
0.1024653466 |
- |
- |
PREDICTED: probable protein phosphatase 2C 49 [Jatropha curcas] |
| 3 |
Hb_001226_170 |
0.1111839449 |
- |
- |
PREDICTED: putative nuclease HARBI1 [Jatropha curcas] |
| 4 |
Hb_002849_110 |
0.1140533892 |
- |
- |
hypothetical protein JCGZ_14362 [Jatropha curcas] |
| 5 |
Hb_007382_100 |
0.11460445 |
- |
- |
receptor serine-threonine protein kinase, putative [Ricinus communis] |
| 6 |
Hb_000836_210 |
0.1166239299 |
- |
- |
hypothetical protein JCGZ_14362 [Jatropha curcas] |
| 7 |
Hb_012787_040 |
0.1280929826 |
- |
- |
PREDICTED: uncharacterized protein LOC105649794 isoform X2 [Jatropha curcas] |
| 8 |
Hb_000172_080 |
0.1326655432 |
transcription factor |
TF Family: ERF |
PREDICTED: ethylene-responsive transcription factor RAP2-4-like [Jatropha curcas] |
| 9 |
Hb_178658_010 |
0.1335021461 |
- |
- |
PREDICTED: uncharacterized protein LOC105641396 [Jatropha curcas] |
| 10 |
Hb_009476_110 |
0.1369449095 |
- |
- |
PREDICTED: probable protein phosphatase 2C 25 [Jatropha curcas] |
| 11 |
Hb_000836_600 |
0.1372044043 |
- |
- |
PREDICTED: delta(8)-fatty-acid desaturase 2 [Jatropha curcas] |
| 12 |
Hb_001619_160 |
0.1417520307 |
transcription factor |
TF Family: ERF |
DREB1p [Hevea brasiliensis] |
| 13 |
Hb_000069_330 |
0.1423000597 |
- |
- |
protein with unknown function [Ricinus communis] |
| 14 |
Hb_000776_050 |
0.1433562424 |
- |
- |
Alpha-1,4-glucan-protein synthase [UDP-forming], putative [Ricinus communis] |
| 15 |
Hb_000805_040 |
0.1447608019 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RING1 [Jatropha curcas] |
| 16 |
Hb_000009_200 |
0.1469609643 |
- |
- |
PREDICTED: UPF0496 protein At2g18630 [Jatropha curcas] |
| 17 |
Hb_098160_010 |
0.1472713941 |
desease resistance |
Gene Name: TIR |
PREDICTED: TMV resistance protein N-like isoform X2 [Jatropha curcas] |
| 18 |
Hb_001329_080 |
0.1489672909 |
- |
- |
PREDICTED: probable protein phosphatase 2C 49 [Jatropha curcas] |
| 19 |
Hb_000373_250 |
0.1514207256 |
- |
- |
PREDICTED: uncharacterized protein LOC105640939 [Jatropha curcas] |
| 20 |
Hb_001102_200 |
0.1514795794 |
- |
- |
PREDICTED: trifunctional UDP-glucose 4,6-dehydratase/UDP-4-keto-6-deoxy-D-glucose 3,5-epimerase/UDP-4-keto-L-rhamnose-reductase RHM1 [Jatropha curcas] |