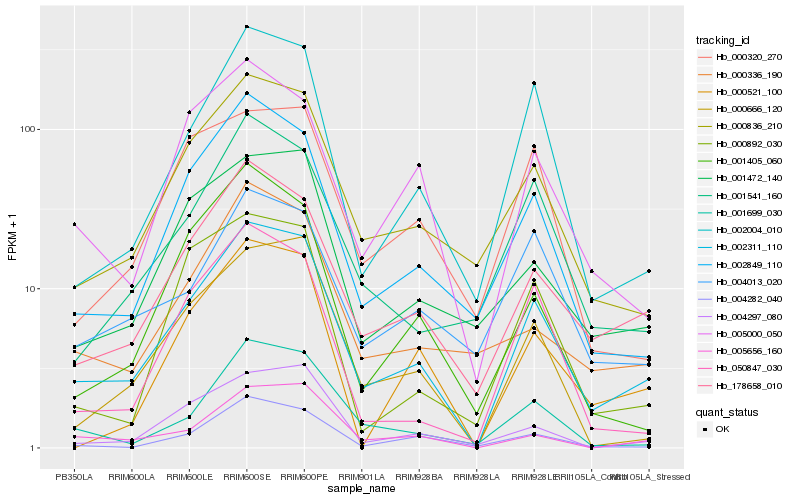
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002311_110 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_178658_010 |
0.1181366663 |
- |
- |
PREDICTED: uncharacterized protein LOC105641396 [Jatropha curcas] |
| 3 |
Hb_000836_210 |
0.1270478315 |
- |
- |
hypothetical protein JCGZ_14362 [Jatropha curcas] |
| 4 |
Hb_002004_010 |
0.1281316956 |
- |
- |
CBL-interacting protein kinase 9 [Populus trichocarpa] |
| 5 |
Hb_002849_110 |
0.1319834408 |
- |
- |
hypothetical protein JCGZ_14362 [Jatropha curcas] |
| 6 |
Hb_000666_120 |
0.1464303091 |
- |
- |
Receptor protein kinase CLAVATA1 precursor, putative [Ricinus communis] |
| 7 |
Hb_005656_160 |
0.1498998306 |
- |
- |
PREDICTED: uncharacterized protein LOC105637448 [Jatropha curcas] |
| 8 |
Hb_001541_160 |
0.1533654565 |
- |
- |
PREDICTED: probable E3 ubiquitin-protein ligase XERICO [Jatropha curcas] |
| 9 |
Hb_001699_030 |
0.1533705449 |
transcription factor |
TF Family: Orphans |
PREDICTED: histidine kinase 1 isoform X1 [Jatropha curcas] |
| 10 |
Hb_001472_140 |
0.1538364617 |
- |
- |
PREDICTED: BTB/POZ domain-containing protein At5g41330 [Jatropha curcas] |
| 11 |
Hb_005000_050 |
0.1565731307 |
- |
- |
PREDICTED: tetraspanin-2 [Jatropha curcas] |
| 12 |
Hb_004297_080 |
0.1574416336 |
- |
- |
PREDICTED: UPF0481 protein At3g47200-like [Jatropha curcas] |
| 13 |
Hb_050847_030 |
0.160904622 |
- |
- |
PREDICTED: cytochrome P450 714A1-like [Populus euphratica] |
| 14 |
Hb_004282_040 |
0.1634400528 |
desease resistance |
Gene Name: TIR |
PREDICTED: TMV resistance protein N-like isoform X2 [Jatropha curcas] |
| 15 |
Hb_000336_190 |
0.1662226113 |
- |
- |
PREDICTED: UPF0481 protein At3g47200-like [Gossypium raimondii] |
| 16 |
Hb_004013_020 |
0.1667716544 |
- |
- |
PREDICTED: F-box protein SKIP2 [Jatropha curcas] |
| 17 |
Hb_000521_100 |
0.1712529558 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_001405_060 |
0.1718853378 |
- |
- |
PREDICTED: CBL-interacting serine/threonine-protein kinase 4-like [Jatropha curcas] |
| 19 |
Hb_000892_030 |
0.1733566244 |
- |
- |
PREDICTED: uncharacterized protein At1g04910 [Jatropha curcas] |
| 20 |
Hb_000320_270 |
0.1742007908 |
- |
- |
PREDICTED: 5'-adenylylsulfate reductase 1, chloroplastic [Jatropha curcas] |