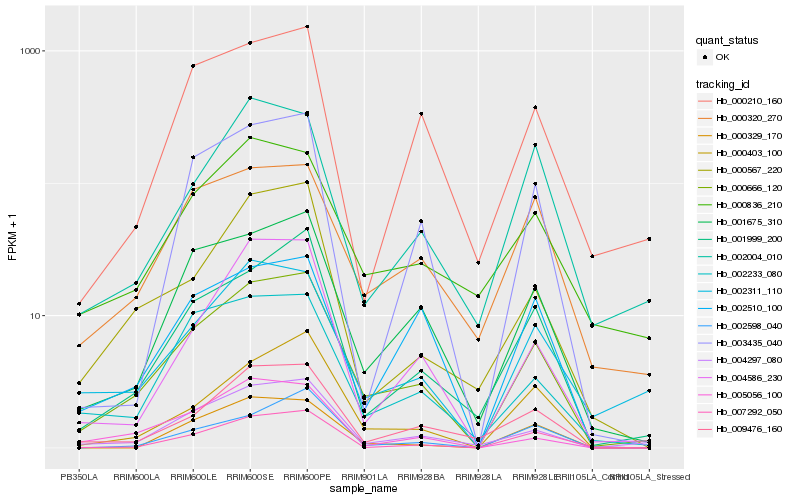
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000666_120 |
0.0 |
- |
- |
Receptor protein kinase CLAVATA1 precursor, putative [Ricinus communis] |
| 2 |
Hb_001675_310 |
0.1214186949 |
transcription factor |
TF Family: AUX/IAA |
PREDICTED: auxin-responsive protein IAA27-like isoform X1 [Jatropha curcas] |
| 3 |
Hb_004297_080 |
0.1232468879 |
- |
- |
PREDICTED: UPF0481 protein At3g47200-like [Jatropha curcas] |
| 4 |
Hb_001999_200 |
0.1374316083 |
transcription factor |
TF Family: C2H2 |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_000403_100 |
0.1376337148 |
- |
- |
PREDICTED: ras-related protein Rab7 [Jatropha curcas] |
| 6 |
Hb_007292_050 |
0.1457454959 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: disease resistance RPP13-like protein 4 [Jatropha curcas] |
| 7 |
Hb_002311_110 |
0.1464303091 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_009476_160 |
0.1468774374 |
- |
- |
RING-H2 finger protein ATL2N, putative [Ricinus communis] |
| 9 |
Hb_002233_080 |
0.1476142203 |
- |
- |
hypothetical protein JCGZ_00222 [Jatropha curcas] |
| 10 |
Hb_000320_270 |
0.1541930738 |
- |
- |
PREDICTED: 5'-adenylylsulfate reductase 1, chloroplastic [Jatropha curcas] |
| 11 |
Hb_004586_230 |
0.1562571826 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_000836_210 |
0.1577526595 |
- |
- |
hypothetical protein JCGZ_14362 [Jatropha curcas] |
| 13 |
Hb_000567_220 |
0.1587499878 |
- |
- |
PREDICTED: uncharacterized protein LOC105116038 [Populus euphratica] |
| 14 |
Hb_000210_160 |
0.1649083908 |
- |
- |
PREDICTED: protein YLS9-like [Jatropha curcas] |
| 15 |
Hb_003435_040 |
0.1659748702 |
- |
- |
tonoplast intrinsic protein [Hevea brasiliensis] |
| 16 |
Hb_002598_040 |
0.1670072313 |
- |
- |
hypothetical protein OsI_22030 [Oryza sativa Indica Group] |
| 17 |
Hb_005056_100 |
0.1672242556 |
transcription factor |
TF Family: AP2 |
PREDICTED: AP2-like ethylene-responsive transcription factor ANT [Jatropha curcas] |
| 18 |
Hb_000329_170 |
0.1680879038 |
- |
- |
PREDICTED: cysteine-rich repeat secretory protein 15 [Jatropha curcas] |
| 19 |
Hb_002510_100 |
0.1689712232 |
- |
- |
Root phototropism protein, putative [Ricinus communis] |
| 20 |
Hb_002004_010 |
0.169819947 |
- |
- |
CBL-interacting protein kinase 9 [Populus trichocarpa] |