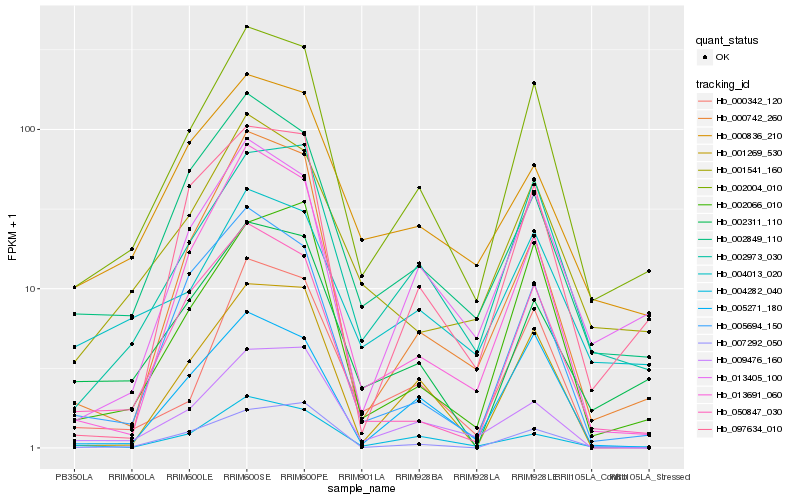
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002004_010 |
0.0 |
- |
- |
CBL-interacting protein kinase 9 [Populus trichocarpa] |
| 2 |
Hb_002973_030 |
0.1028472351 |
- |
- |
CBL-interacting serine/threonine-protein kinase, putative [Ricinus communis] |
| 3 |
Hb_013405_100 |
0.1229287283 |
transcription factor |
TF Family: GRAS |
PREDICTED: scarecrow-like protein 8 [Jatropha curcas] |
| 4 |
Hb_001541_160 |
0.1242583976 |
- |
- |
PREDICTED: probable E3 ubiquitin-protein ligase XERICO [Jatropha curcas] |
| 5 |
Hb_050847_030 |
0.1271358234 |
- |
- |
PREDICTED: cytochrome P450 714A1-like [Populus euphratica] |
| 6 |
Hb_002311_110 |
0.1281316956 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_002849_110 |
0.1284954947 |
- |
- |
hypothetical protein JCGZ_14362 [Jatropha curcas] |
| 8 |
Hb_002066_010 |
0.1346190794 |
- |
- |
PREDICTED: acetylglutamate kinase, chloroplastic [Jatropha curcas] |
| 9 |
Hb_013691_060 |
0.1360896962 |
- |
- |
PREDICTED: uncharacterized protein LOC105119066 [Populus euphratica] |
| 10 |
Hb_004282_040 |
0.1373826467 |
desease resistance |
Gene Name: TIR |
PREDICTED: TMV resistance protein N-like isoform X2 [Jatropha curcas] |
| 11 |
Hb_097634_010 |
0.1402050685 |
- |
- |
PREDICTED: UDP-glucuronate 4-epimerase 1 [Jatropha curcas] |
| 12 |
Hb_000742_260 |
0.1404341766 |
- |
- |
hypothetical protein RCOM_1336320 [Ricinus communis] |
| 13 |
Hb_005271_180 |
0.1416021068 |
- |
- |
Leucine-rich repeat receptor protein kinase EXS precursor, putative [Ricinus communis] |
| 14 |
Hb_000836_210 |
0.1452952538 |
- |
- |
hypothetical protein JCGZ_14362 [Jatropha curcas] |
| 15 |
Hb_009476_160 |
0.1455768418 |
- |
- |
RING-H2 finger protein ATL2N, putative [Ricinus communis] |
| 16 |
Hb_005694_150 |
0.1461247473 |
- |
- |
PREDICTED: probable mitochondrial chaperone BCS1-A [Jatropha curcas] |
| 17 |
Hb_001269_530 |
0.1465706365 |
- |
- |
ATP-binding cassette transporter, putative [Ricinus communis] |
| 18 |
Hb_000342_120 |
0.1479953012 |
- |
- |
PREDICTED: probable protein phosphatase 2C 72 [Jatropha curcas] |
| 19 |
Hb_004013_020 |
0.1528516337 |
- |
- |
PREDICTED: F-box protein SKIP2 [Jatropha curcas] |
| 20 |
Hb_007292_050 |
0.1560349194 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: disease resistance RPP13-like protein 4 [Jatropha curcas] |