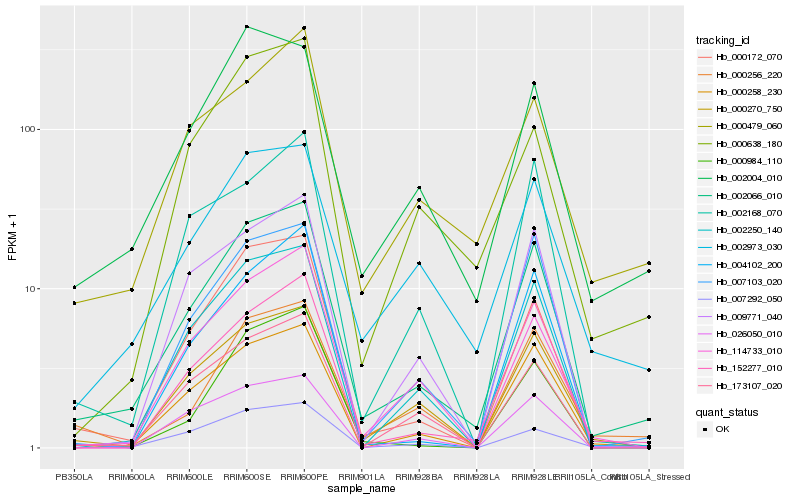
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002066_010 |
0.0 |
- |
- |
PREDICTED: acetylglutamate kinase, chloroplastic [Jatropha curcas] |
| 2 |
Hb_000258_230 |
0.1154237797 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_000172_070 |
0.1175232625 |
- |
- |
trehalose-6-phosphate synthase, putative [Ricinus communis] |
| 4 |
Hb_152277_010 |
0.1239081362 |
- |
- |
PREDICTED: 9-cis-epoxycarotenoid dioxygenase NCED3, chloroplastic-like [Jatropha curcas] |
| 5 |
Hb_007103_020 |
0.1245545144 |
- |
- |
hypothetical protein POPTR_0007s04191g [Populus trichocarpa] |
| 6 |
Hb_002250_140 |
0.1245654329 |
- |
- |
PREDICTED: EPIDERMAL PATTERNING FACTOR-like protein 6 isoform X1 [Populus euphratica] |
| 7 |
Hb_000270_750 |
0.1279351855 |
- |
- |
PREDICTED: RING-H2 finger protein ATL46-like [Populus euphratica] |
| 8 |
Hb_007292_050 |
0.1286687368 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: disease resistance RPP13-like protein 4 [Jatropha curcas] |
| 9 |
Hb_002973_030 |
0.1308047727 |
- |
- |
CBL-interacting serine/threonine-protein kinase, putative [Ricinus communis] |
| 10 |
Hb_002004_010 |
0.1346190794 |
- |
- |
CBL-interacting protein kinase 9 [Populus trichocarpa] |
| 11 |
Hb_173107_020 |
0.1373797897 |
- |
- |
PREDICTED: putative receptor-like protein kinase At1g80870 [Jatropha curcas] |
| 12 |
Hb_000256_220 |
0.1378265553 |
- |
- |
PREDICTED: flotillin-like protein 4 [Jatropha curcas] |
| 13 |
Hb_009771_040 |
0.1386471275 |
- |
- |
hypothetical protein JCGZ_25039 [Jatropha curcas] |
| 14 |
Hb_114733_010 |
0.138918419 |
- |
- |
PREDICTED: anthocyanidin 5,3-O-glucosyltransferase-like [Jatropha curcas] |
| 15 |
Hb_000479_060 |
0.1431756671 |
- |
- |
PREDICTED: UDP-glucose 6-dehydrogenase 1 [Vitis vinifera] |
| 16 |
Hb_000984_110 |
0.1437381444 |
- |
- |
hypothetical protein JCGZ_15953 [Jatropha curcas] |
| 17 |
Hb_000638_180 |
0.1456886235 |
- |
- |
PREDICTED: uncharacterized protein LOC105124685 [Populus euphratica] |
| 18 |
Hb_026050_010 |
0.1463051343 |
- |
- |
protein phosphatase 2c, putative [Ricinus communis] |
| 19 |
Hb_002168_070 |
0.1465145857 |
- |
- |
Jasmonate O-methyltransferase, putative [Ricinus communis] |
| 20 |
Hb_004102_200 |
0.14872857 |
- |
- |
PREDICTED: putative UDP-rhamnose:rhamnosyltransferase 1 [Jatropha curcas] |