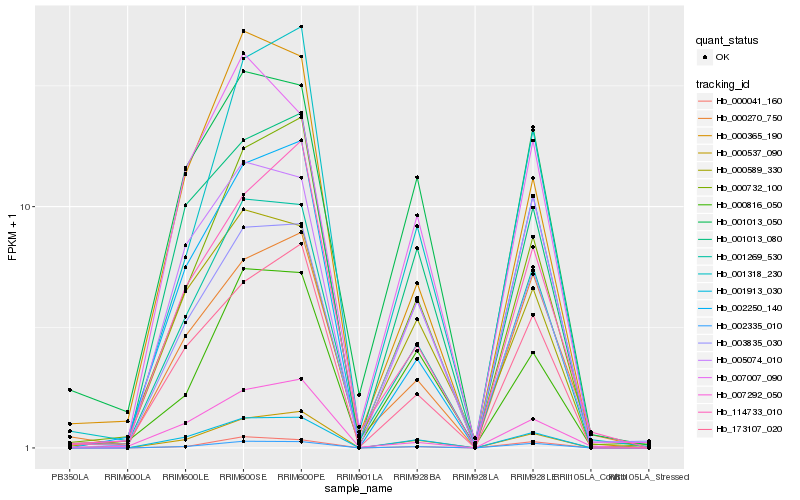
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001269_530 |
0.0 |
- |
- |
ATP-binding cassette transporter, putative [Ricinus communis] |
| 2 |
Hb_001913_030 |
0.0696730343 |
- |
- |
ankyrin repeat protein [Bruguiera gymnorhiza] |
| 3 |
Hb_000537_090 |
0.0869022433 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_002335_010 |
0.0900435209 |
- |
- |
hypothetical protein JCGZ_23475 [Jatropha curcas] |
| 5 |
Hb_000732_100 |
0.0965982443 |
- |
- |
PREDICTED: uncharacterized protein LOC105649440 [Jatropha curcas] |
| 6 |
Hb_000589_330 |
0.0989019852 |
- |
- |
kinase, putative [Ricinus communis] |
| 7 |
Hb_173107_020 |
0.1013578505 |
- |
- |
PREDICTED: putative receptor-like protein kinase At1g80870 [Jatropha curcas] |
| 8 |
Hb_005074_010 |
0.1073491071 |
- |
- |
PREDICTED: uncharacterized protein LOC105644456 [Jatropha curcas] |
| 9 |
Hb_002250_140 |
0.1077248314 |
- |
- |
PREDICTED: EPIDERMAL PATTERNING FACTOR-like protein 6 isoform X1 [Populus euphratica] |
| 10 |
Hb_001013_050 |
0.1080878372 |
- |
- |
unnamed protein product [Coffea canephora] |
| 11 |
Hb_001013_080 |
0.1105927904 |
- |
- |
anthocyanidin 3-O-glucosyltransferase [Hevea brasiliensis] |
| 12 |
Hb_003835_030 |
0.1122695898 |
- |
- |
glycosyltransferase, partial [Arabidopsis thaliana] |
| 13 |
Hb_001318_230 |
0.1135515066 |
- |
- |
Boron transporter, putative [Ricinus communis] |
| 14 |
Hb_007007_090 |
0.1138687069 |
- |
- |
phospholipase d beta, putative [Ricinus communis] |
| 15 |
Hb_000365_190 |
0.1159608405 |
- |
- |
PREDICTED: protein LURP-one-related 8 [Jatropha curcas] |
| 16 |
Hb_000270_750 |
0.1161526734 |
- |
- |
PREDICTED: RING-H2 finger protein ATL46-like [Populus euphratica] |
| 17 |
Hb_000041_160 |
0.1230714344 |
- |
- |
PREDICTED: uncharacterized protein LOC104907627 [Beta vulgaris subsp. vulgaris] |
| 18 |
Hb_114733_010 |
0.1242150877 |
- |
- |
PREDICTED: anthocyanidin 5,3-O-glucosyltransferase-like [Jatropha curcas] |
| 19 |
Hb_000816_050 |
0.125849057 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: disease resistance protein RGA2-like [Jatropha curcas] |
| 20 |
Hb_007292_050 |
0.1277445181 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: disease resistance RPP13-like protein 4 [Jatropha curcas] |