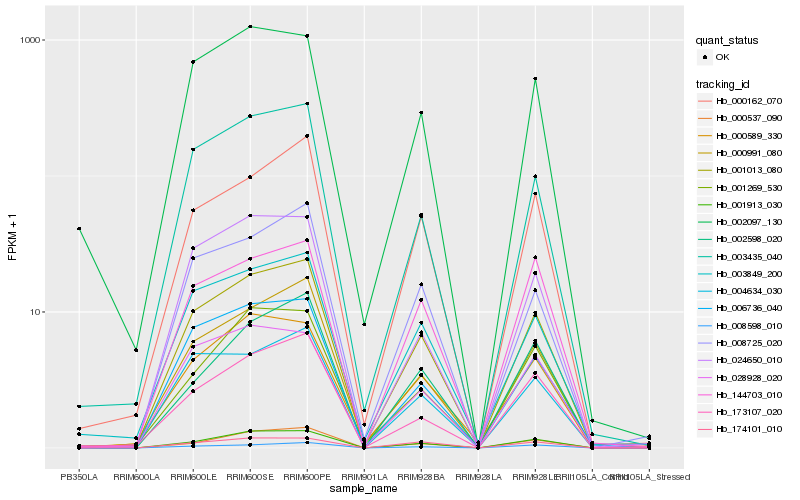
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001013_080 |
0.0 |
- |
- |
anthocyanidin 3-O-glucosyltransferase [Hevea brasiliensis] |
| 2 |
Hb_001913_030 |
0.0686138051 |
- |
- |
ankyrin repeat protein [Bruguiera gymnorhiza] |
| 3 |
Hb_000991_080 |
0.0726596696 |
- |
- |
kinase, putative [Ricinus communis] |
| 4 |
Hb_003435_040 |
0.0798867166 |
- |
- |
tonoplast intrinsic protein [Hevea brasiliensis] |
| 5 |
Hb_003849_200 |
0.0823686746 |
- |
- |
PREDICTED: BTB/POZ domain-containing protein At1g67900-like isoform X1 [Citrus sinensis] |
| 6 |
Hb_000162_070 |
0.0870020012 |
- |
- |
phospholipase C, putative [Ricinus communis] |
| 7 |
Hb_008725_020 |
0.0897347354 |
- |
- |
PREDICTED: cytidine deaminase 1 [Jatropha curcas] |
| 8 |
Hb_006736_040 |
0.0937510927 |
- |
- |
PREDICTED: glucan endo-1,3-beta-glucosidase, basic isoform-like [Populus euphratica] |
| 9 |
Hb_000537_090 |
0.0939304656 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_008598_010 |
0.0967966031 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase At1g61370 [Jatropha curcas] |
| 11 |
Hb_004634_030 |
0.0974037772 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_000589_330 |
0.1004435535 |
- |
- |
kinase, putative [Ricinus communis] |
| 13 |
Hb_173107_020 |
0.1045028541 |
- |
- |
PREDICTED: putative receptor-like protein kinase At1g80870 [Jatropha curcas] |
| 14 |
Hb_001269_530 |
0.1105927904 |
- |
- |
ATP-binding cassette transporter, putative [Ricinus communis] |
| 15 |
Hb_024650_010 |
0.1113791644 |
- |
- |
PREDICTED: BAHD acyltransferase DCR [Jatropha curcas] |
| 16 |
Hb_028928_020 |
0.1131875342 |
- |
- |
hypothetical protein CICLE_v10018504mg [Citrus clementina] |
| 17 |
Hb_002097_130 |
0.1143231376 |
- |
- |
tonoplast intrinsic protein [Hevea brasiliensis] |
| 18 |
Hb_144703_010 |
0.1148227184 |
- |
- |
Phospholipase A 2A, IIA,PLA2A [Theobroma cacao] |
| 19 |
Hb_174101_010 |
0.1180759322 |
- |
- |
PREDICTED: receptor-like protein 12 [Populus euphratica] |
| 20 |
Hb_002598_020 |
0.1182764814 |
- |
- |
PREDICTED: sugar transporter ERD6-like 5 [Populus euphratica] |