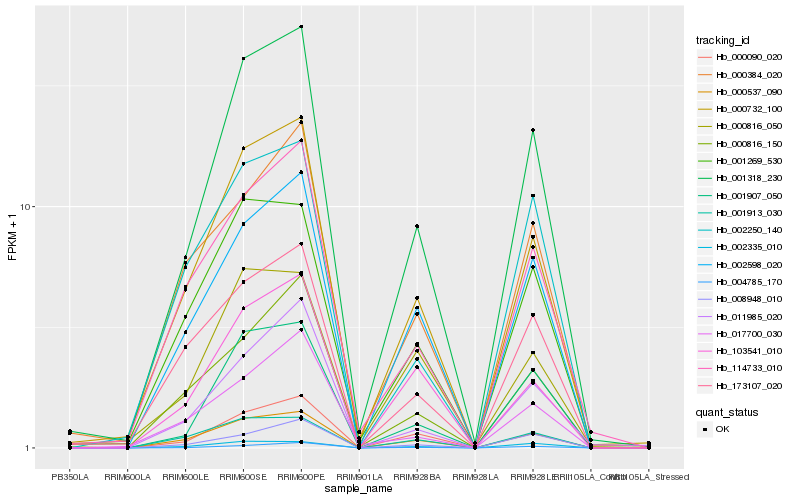
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001318_230 |
0.0 |
- |
- |
Boron transporter, putative [Ricinus communis] |
| 2 |
Hb_000732_100 |
0.0676601986 |
- |
- |
PREDICTED: uncharacterized protein LOC105649440 [Jatropha curcas] |
| 3 |
Hb_001907_050 |
0.0852927685 |
- |
- |
Receptor like protein 6, putative [Theobroma cacao] |
| 4 |
Hb_000537_090 |
0.0898121608 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_002598_020 |
0.1039194486 |
- |
- |
PREDICTED: sugar transporter ERD6-like 5 [Populus euphratica] |
| 6 |
Hb_000090_020 |
0.106348195 |
- |
- |
acyltransferase, putative [Ricinus communis] |
| 7 |
Hb_004785_170 |
0.1107046375 |
- |
- |
PREDICTED: receptor-like protein 12 [Jatropha curcas] |
| 8 |
Hb_114733_010 |
0.1121282932 |
- |
- |
PREDICTED: anthocyanidin 5,3-O-glucosyltransferase-like [Jatropha curcas] |
| 9 |
Hb_001269_530 |
0.1135515066 |
- |
- |
ATP-binding cassette transporter, putative [Ricinus communis] |
| 10 |
Hb_011985_020 |
0.1251747288 |
- |
- |
PREDICTED: phospholipase A1-Igamma3, chloroplastic [Jatropha curcas] |
| 11 |
Hb_000816_150 |
0.1256129034 |
- |
- |
oligopeptide transporter, putative [Ricinus communis] |
| 12 |
Hb_173107_020 |
0.126543838 |
- |
- |
PREDICTED: putative receptor-like protein kinase At1g80870 [Jatropha curcas] |
| 13 |
Hb_008948_010 |
0.1267523528 |
- |
- |
PREDICTED: equilibrative nucleotide transporter 8 isoform X1 [Jatropha curcas] |
| 14 |
Hb_002335_010 |
0.1295634005 |
- |
- |
hypothetical protein JCGZ_23475 [Jatropha curcas] |
| 15 |
Hb_103541_010 |
0.1309863357 |
- |
- |
PREDICTED: probable leucine-rich repeat receptor-like protein kinase At5g63930 isoform X2 [Jatropha curcas] |
| 16 |
Hb_000816_050 |
0.1331336065 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: disease resistance protein RGA2-like [Jatropha curcas] |
| 17 |
Hb_000384_020 |
0.1363780697 |
- |
- |
sucrose phosphate syntase, putative [Ricinus communis] |
| 18 |
Hb_001913_030 |
0.1383793454 |
- |
- |
ankyrin repeat protein [Bruguiera gymnorhiza] |
| 19 |
Hb_002250_140 |
0.1388973198 |
- |
- |
PREDICTED: EPIDERMAL PATTERNING FACTOR-like protein 6 isoform X1 [Populus euphratica] |
| 20 |
Hb_017700_030 |
0.1395215007 |
- |
- |
PREDICTED: vacuolar amino acid transporter 1 [Jatropha curcas] |