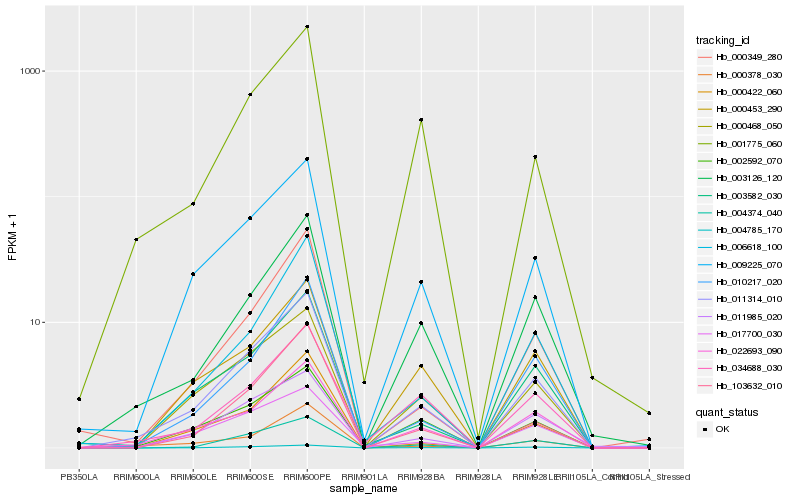
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_011314_010 |
0.0 |
- |
- |
PREDICTED: disease resistance protein RPS6-like isoform X1 [Jatropha curcas] |
| 2 |
Hb_009225_070 |
0.0860002113 |
- |
- |
tonoplast intrinsic protein, putative [Ricinus communis] |
| 3 |
Hb_022693_090 |
0.0982784085 |
- |
- |
glycosyltransferase, putative [Ricinus communis] |
| 4 |
Hb_004374_040 |
0.1053751897 |
- |
- |
PREDICTED: beta-carotene 3-hydroxylase, chloroplastic-like [Jatropha curcas] |
| 5 |
Hb_003126_120 |
0.1073789758 |
- |
- |
PREDICTED: cytochrome b561 and DOMON domain-containing protein At5g47530-like [Jatropha curcas] |
| 6 |
Hb_004785_170 |
0.1099434098 |
- |
- |
PREDICTED: receptor-like protein 12 [Jatropha curcas] |
| 7 |
Hb_003582_030 |
0.1102828784 |
- |
- |
PREDICTED: serine/threonine-protein kinase BRI1-like 2 [Jatropha curcas] |
| 8 |
Hb_000468_050 |
0.1127331711 |
- |
- |
- |
| 9 |
Hb_000378_030 |
0.1158849682 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_010217_020 |
0.1187897102 |
- |
- |
cinnamate 4-hydroxylase [Leucaena leucocephala] |
| 11 |
Hb_002592_070 |
0.1206654931 |
- |
- |
hypothetical protein VITISV_020993 [Vitis vinifera] |
| 12 |
Hb_000349_280 |
0.1268589544 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_103632_010 |
0.1275775234 |
- |
- |
triacylglycerol lipase, putative [Ricinus communis] |
| 14 |
Hb_000422_060 |
0.1292439415 |
transcription factor |
TF Family: OFP |
hypothetical protein RCOM_1282480 [Ricinus communis] |
| 15 |
Hb_011985_020 |
0.1305119873 |
- |
- |
PREDICTED: phospholipase A1-Igamma3, chloroplastic [Jatropha curcas] |
| 16 |
Hb_017700_030 |
0.1305977327 |
- |
- |
PREDICTED: vacuolar amino acid transporter 1 [Jatropha curcas] |
| 17 |
Hb_000453_290 |
0.1327770118 |
- |
- |
PREDICTED: dr1-associated corepressor-like [Jatropha curcas] |
| 18 |
Hb_034688_030 |
0.1330381718 |
- |
- |
PREDICTED: uncharacterized protein LOC105642049 [Jatropha curcas] |
| 19 |
Hb_006618_100 |
0.13335162 |
transcription factor |
TF Family: MYB |
MYB transcriptional factor [Populus tremula x Populus tremuloides] |
| 20 |
Hb_001775_060 |
0.1340693517 |
- |
- |
conserved hypothetical protein [Ricinus communis] |