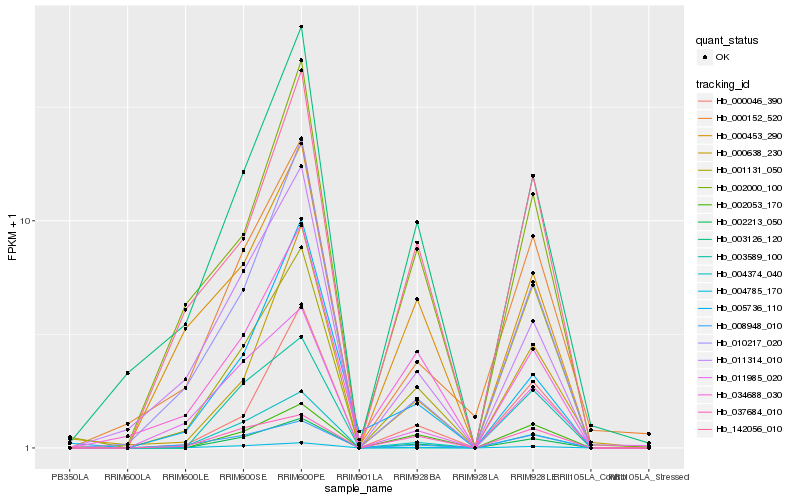
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002213_050 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105630318 isoform X3 [Jatropha curcas] |
| 2 |
Hb_004374_040 |
0.1092431136 |
- |
- |
PREDICTED: beta-carotene 3-hydroxylase, chloroplastic-like [Jatropha curcas] |
| 3 |
Hb_002053_170 |
0.1253923858 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 4 |
Hb_004785_170 |
0.1286789375 |
- |
- |
PREDICTED: receptor-like protein 12 [Jatropha curcas] |
| 5 |
Hb_003126_120 |
0.1535496274 |
- |
- |
PREDICTED: cytochrome b561 and DOMON domain-containing protein At5g47530-like [Jatropha curcas] |
| 6 |
Hb_000046_390 |
0.1589355981 |
- |
- |
PREDICTED: CASP-like protein 1D1 [Jatropha curcas] |
| 7 |
Hb_000152_520 |
0.1696479278 |
transcription factor |
TF Family: MYB |
hypothetical protein POPTR_0015s14600g [Populus trichocarpa] |
| 8 |
Hb_034688_030 |
0.1739827217 |
- |
- |
PREDICTED: uncharacterized protein LOC105642049 [Jatropha curcas] |
| 9 |
Hb_011314_010 |
0.1740996798 |
- |
- |
PREDICTED: disease resistance protein RPS6-like isoform X1 [Jatropha curcas] |
| 10 |
Hb_037684_010 |
0.1744961052 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC105645015 [Jatropha curcas] |
| 11 |
Hb_000638_230 |
0.177811308 |
transcription factor |
TF Family: MYB |
r2r3-myb transcription factor, putative [Ricinus communis] |
| 12 |
Hb_008948_010 |
0.1783595727 |
- |
- |
PREDICTED: equilibrative nucleotide transporter 8 isoform X1 [Jatropha curcas] |
| 13 |
Hb_010217_020 |
0.1787884752 |
- |
- |
cinnamate 4-hydroxylase [Leucaena leucocephala] |
| 14 |
Hb_002000_100 |
0.1862558904 |
- |
- |
PREDICTED: cellulose synthase A catalytic subunit 2 [UDP-forming]-like [Jatropha curcas] |
| 15 |
Hb_142056_010 |
0.1863859543 |
- |
- |
PREDICTED: MATE efflux family protein 1 isoform X2 [Jatropha curcas] |
| 16 |
Hb_001131_050 |
0.1864766649 |
- |
- |
hypothetical protein EUGRSUZ_C02968 [Eucalyptus grandis] |
| 17 |
Hb_011985_020 |
0.1885379714 |
- |
- |
PREDICTED: phospholipase A1-Igamma3, chloroplastic [Jatropha curcas] |
| 18 |
Hb_005736_110 |
0.1912723067 |
transcription factor |
TF Family: MYB |
MYB transcriptional factor [Populus tremula x Populus tremuloides] |
| 19 |
Hb_003589_100 |
0.1927213042 |
- |
- |
hypothetical protein JCGZ_03338 [Jatropha curcas] |
| 20 |
Hb_000453_290 |
0.1929602131 |
- |
- |
PREDICTED: dr1-associated corepressor-like [Jatropha curcas] |