| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
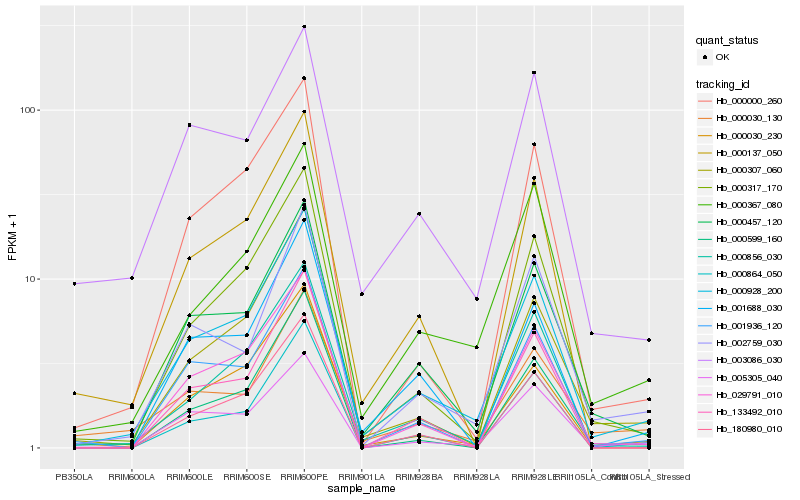
Hb_000928_200 |
0.0 |
- |
- |
PREDICTED: RING-H2 finger protein ATL52-like [Jatropha curcas] |
| 2 |
Hb_000457_120 |
0.0849183247 |
- |
- |
PREDICTED: death-associated protein kinase dapk-1 [Jatropha curcas] |
| 3 |
Hb_000317_170 |
0.0923796775 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RING1 [Jatropha curcas] |
| 4 |
Hb_005305_040 |
0.1006341483 |
transcription factor |
TF Family: C3H |
transcription factor, putative [Ricinus communis] |
| 5 |
Hb_000856_030 |
0.1028929259 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 6 |
Hb_000030_230 |
0.104420698 |
- |
- |
PREDICTED: pyruvate kinase, cytosolic isozyme [Jatropha curcas] |
| 7 |
Hb_000307_060 |
0.1066842498 |
- |
- |
PREDICTED: mechanosensitive ion channel protein 8-like [Jatropha curcas] |
| 8 |
Hb_000367_080 |
0.1077496308 |
- |
- |
PREDICTED: UDP-glucose 6-dehydrogenase 1-like [Jatropha curcas] |
| 9 |
Hb_002759_030 |
0.1116187838 |
- |
- |
PREDICTED: serine/threonine-protein kinase PBS1 [Jatropha curcas] |
| 10 |
Hb_000000_260 |
0.1118188692 |
- |
- |
PREDICTED: probable galacturonosyltransferase-like 1 [Jatropha curcas] |
| 11 |
Hb_029791_010 |
0.1136215928 |
- |
- |
hypothetical protein POPTR_0014s17480g [Populus trichocarpa] |
| 12 |
Hb_001936_120 |
0.114600474 |
- |
- |
PREDICTED: uncharacterized protein LOC105644623 isoform X1 [Jatropha curcas] |
| 13 |
Hb_000137_050 |
0.1161373881 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_000864_050 |
0.1218570037 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 15 |
Hb_000030_130 |
0.1247443085 |
transcription factor |
TF Family: G2-like |
transcription factor, putative [Ricinus communis] |
| 16 |
Hb_001688_030 |
0.1251630365 |
- |
- |
hypothetical protein JCGZ_10716 [Jatropha curcas] |
| 17 |
Hb_000599_160 |
0.1271871619 |
transcription factor |
TF Family: NAC |
PREDICTED: NAC domain-containing protein 21/22 [Jatropha curcas] |
| 18 |
Hb_003086_030 |
0.1287028419 |
- |
- |
PREDICTED: fasciclin-like arabinogalactan protein 17 [Jatropha curcas] |
| 19 |
Hb_180980_010 |
0.1317206673 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At3g47570 [Malus domestica] |
| 20 |
Hb_133492_010 |
0.1332282249 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |